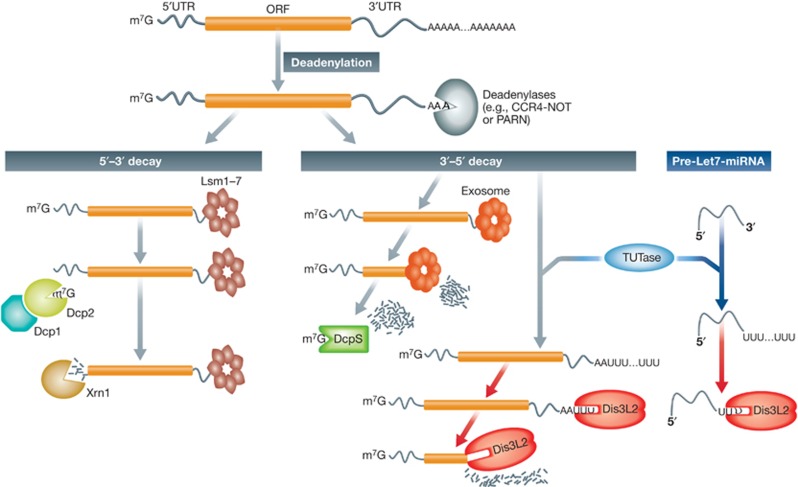
Figure 1.
Dis3L2 is a new player in a major pathway of RNA decay. Many mRNAs normally undergo poly(A) tail removal by deadenylases as a first step prior to their decay by an exonucleolytic mechanism. Recruitment of the Lsm1-7 complex to the 3′ end of a mRNA triggers decapping by the Dcp1–Dcp2 complex, making the RNA accessible to the 5′–3′ exoribonuclease Xrn1. Degradation can also occur in the 3′→5′ direction if the exosome complex is recruited to the 3′ end of deadenylated transcript. In this pathway, short-capped RNA oligomers generated by the exosome are acted on by the scavenger-decapping enzyme DcpS. The recent observations of Malecki et al (2013) and Lubas et al (2013) indicate that the 3′–5′ decay of mRNAs can also occur in an exosome-independent and uridylation-dependent manner. A TUTase enzyme adds a short poly(U) tract to a deadenylated mRNA that becomes an ideal landing pad for the 3′–5′ exoribonuclease Dis3L2. Chang et al (2013) demonstrated that a similar uridylation/Dis3L2-mediated 3′–5′ decay pathway also acts on let7 pre-miRNAs.