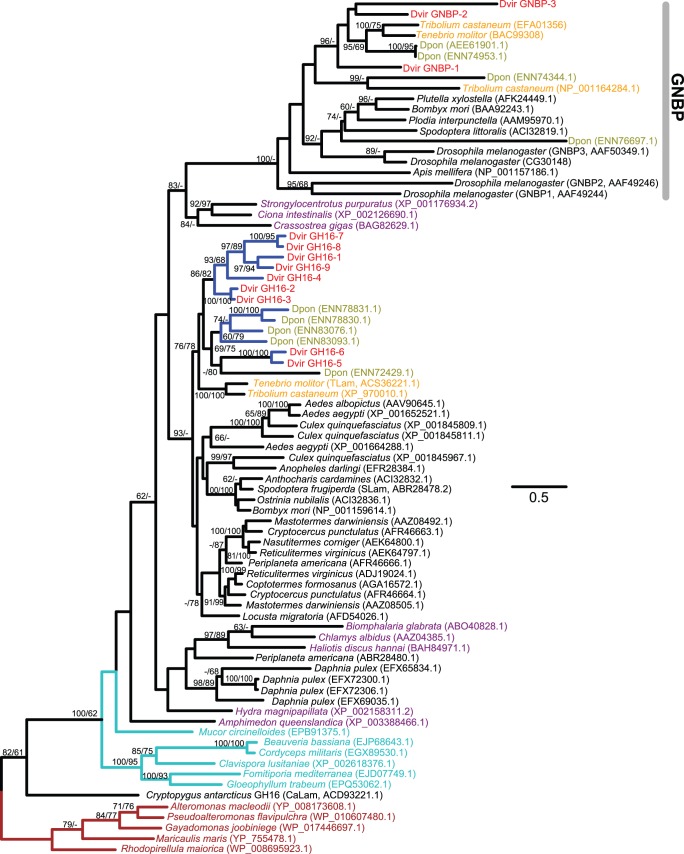
Figure 5. The maximum-likelihood phylogeny of GH16 proteins.
Sixteen GH16 protein sequences from four coleopteran species are included. Labels for the coleopteran species belonging to the superfamily Curculionoidea, D. v. virgifera, and other beetle sequences are shown in olive, red, and orange, respectively. Their species abbreviations are found in Table S5. Arthropod, other metazoan, fungal (6 chosen from 222 sequences), and bacterial (5 chosen from 977 sequences) sequences are indicated by black, purple, cyan, and brown, respectively. Bacterial sequences were used as outgroups. The numbers at internal branches show the bootstrap support values (%) for the maximum-likelihood and neighbor-joining phylogenies in this order. Supporting values are shown only when higher than 60%. Blue-colored branches indicate the species-specific gene duplications (based on currently available sequences) within a cluster supported by higher than 70% of bootstrap values. The scale bar represents the number of amino acid substitutions per site.