Abstract
The phase transition from vegetative to reproductive growth is a critical event in the life cycle of flowering plants. FLOWERING LOCUS T (FT) plays a central role in the regulation of this transition by integrating signals from multiple flowering pathways in the leaves and transmitting them to the shoot apical meristem. In this study, we characterized FT homologs in the temperate grasses Brachypodium distachyon and polyploid wheat using transgenic and mutant approaches. Downregulation of FT1 by RNAi was associated with a significant downregulation of the FT-like genes FT2 and FT4 in Brachypodium and FT2 and FT5 in wheat. In a transgenic wheat line carrying a highly-expressed FT1 allele, FT2 and FT3 were upregulated under both long and short days. Overexpression of FT1 caused extremely early flowering during shoot regeneration in both Brachypodium and hexaploid wheat, and resulted in insufficient vegetative tissue to support the production of viable seeds. Downregulation of FT1 transcripts by RNA interference (RNAi) resulted in non-flowering Brachypodium plants and late flowering plants (2–4 weeks delay) in wheat. A similar delay in heading time was observed in tetraploid wheat plants carrying mutations for both FT-A1 and FT-B1. Plants homozygous only for mutations in FT-B1 flowered later than plants homozygous only for mutations in FT-A1, which corresponded with higher transcript levels of FT-B1 relative to FT-A1 in the early stages of development. Taken together, our data indicate that FT1 plays a critical role in the regulation of flowering in Brachypodium and wheat, and that this role is associated with the simultaneous regulation of other FT-like genes. The differential effects of mutations in FT-A1 and FT-B1 on wheat heading time suggest that different allelic combinations of FT1 homoeologs could be used to adjust wheat heading time to improve adaptation to changing environments.
Introduction
The optimization of flowering and seed production is critical for plant survival and, in seed crops, to maximize grain yields. This is particularly important for cereal crops, which contribute significantly to global food production. However, the regulatory gene network controlling flowering is complex and its manipulation requires a precise understanding of the roles of different components and their interactions. In the temperate grasses, which include wheat, barley and the model species Brachypodium distachyon, vernalization (long exposures to cold temperatures) and photoperiod (variation in day length) are the main seasonal signals regulating flowering time. Over the last two decades, substantial progress has been made toward our understanding of the genes involved in these two pathways [1]–[3].
Vernalization requirement is conferred by the genes VERNALIZATION1 (VRN1) [4], [5], VRN2 [6], VRN3 [7], and VRN4 [8], [9]. Among these four genes, only the first three have been cloned and characterized to date. VRN1 is a MADS-box meristem identity gene homologous to Arabidopsis AP1, which is upregulated during vernalization and promotes flowering in the spring [4]. VRN2 is a floral repressor that encodes a protein containing a zinc finger motif and a CCT domain (CONSTANS, CONSTANS-LIKE, and TIMING OF CAB1-1) [6], and is downregulated by vernalization and short days (SD) [10], [11]. The VRN3 gene encodes a RAF kinase inhibitor-like protein with high similarity to Arabidopsis protein FLOWERING LOCUS T (FT) and is a flowering promoter [7]. This gene will be referred to hereafter as FT1 to differentiate it from other FT-like genes present in the temperate grasses ([12]).
These three vernalization genes are interconnected by complex interactions [3]. VRN2 acts as a repressor of FT1 in the leaves, preventing flowering in the fall. During the winter, FT1 and VRN2 transcripts are reduced to almost undetectable levels whereas VRN1 transcripts increase proportionally to the duration of the cold period [7]. At the end of the winter, the presence of VRN1 represses VRN2 facilitating the upregulation of FT1 during the lengthening days of spring [13]. FT1 expression then further upregulates VRN1 to reinforcing this inductive pathway and result in an irreversible transition to flowering.
Under long days (LD), FT1 is upregulated by the photoperiod gene PHOTOPERIOD 1 (PPD1), which is responsible for most of the natural variation in photoperiod sensitivity in wheat and barley [14]–[16]. The convergence of the photoperiod and vernalization pathways in the regulation of FT1, place this gene at the center of the gene network regulating flowering time in the temperate cereals [3].
In addition to its role in the integration of flowering signals, the small proteins encoded by FT and FT-like genes are essential for the transmission of environmental signals from the leaves to the shoot apical meristem, as demonstrated in Arabidopsis, rice and other species [17]–[20]. FT1 protein transport has yet to be demonstrated in the temperate cereals, but indirect evidence suggests that a similar mechanism exists in these species. FT1 is known to interact with the bZIP transcription factor FDL2, which binds to the promoter of VRN1 [21]. Although FT1 is not expressed in the wheat apex [22], a good correlation has been observed between the transcript levels of FT1 in the leaves and of VRN1 in the apex [21]. A significant correlation between flowering and FT1 transcript levels in the leaves has also been observed in a sample of Brachypodium accessions [23].
Previous studies have shown that over-expression of FT1 accelerates flowering in Brachypodium and wheat [24]–[26] but the effects of the downregulation of this gene in these species have not been reported before. In this study, we validate the FT1 over-expression results and determine the effect of the downregulation of this gene by RNA interference (RNAi) in Brachypodium and wheat. We also use these transgenic lines to characterize the expression of the closest FT-like genes (FT2 to FT6). Finally, we test the effect of mutations in FT1 homoeologs in tetraploid wheat to determine their individual and combined contribution to heading time, and discuss their potential value for engineering wheat flowering time.
Materials and Methods
The materials used in this study included the diploid Brachypodium distachyon (accession ‘Bd21-3′), diploid wheat Triticum monococcum (accession ‘G3116’), tetraploid wheat (T. turgidum ssp. durum cultivars ‘Kronos’ and ‘Langdon’), hexaploid wheat (T. aestivum cultivars ‘Bobwhite’ and ‘Jagger’), and a transgenic Jagger line carrying a highly expressed FT-B1 allele from the variety Hope (referred to hereafter as FT1 HOPE) driven by its native promoter [7].
Brachypodium plants were grown in a growth chamber at 25°C and LD photoperiod (16 h light/8 h dark, light intensity of 36 μmol m−2 s−1). Wild type Bobwhite plants used for transformation were grown in the field (University farm at Tai’an, Shandong, China). Transgenic plants for FT1 silencing using RNA interference (FT1 RNAi) were evaluated in greenhouses in China at 25°C and long days (16 h light/8 h dark, light intensity of 105 μmol m−2 s−1). Wheat mutants were evaluated in greenhouses in California under LD conditions (16 h light) with temperatures that oscillated between 21 and 23°C during the day and between 12 and 18°C during the night. FT1 HOPE overexpressing lines were evaluated in Conviron CMP3244 growth chambers (Conviron, Pembina, ND, USA) under LD (16 h light, 6∶00 a.m. –10∶00 p.m., 16°C) or SD (8 h light, 6∶00 a.m. –2∶00 p.m., 16°C).
Plasmid Construction
To characterize the function of FT1 genes, full-length FT1 cDNAs were cloned from Brachypodium Bd21-3 (GT847109), T. monococcum G3116 (FT-Am1, DQ890163), and tetraploid wheat cultivar Langdon (FT-B1, DQ890164). The genomic region spanning the start to stop codons of the FT-B1 gene was also cloned from Langdon (DQ890164).
The FT1 overexpression (FT1 OE) constructs were developed in binary vectors pCAMBIA1300 and pGWB5 (Figure 1A–B) [27]. The pCAMBIA1300 vector was used for the overexpression of FT1 cDNAs of Bd21-3 and Langdon, and the FT1 genomic region of Langdon. The pGWB5 vector was used for the overexpression of a Brachypodium FT1 cDNA fused to the green fluorescent protein (GFP).
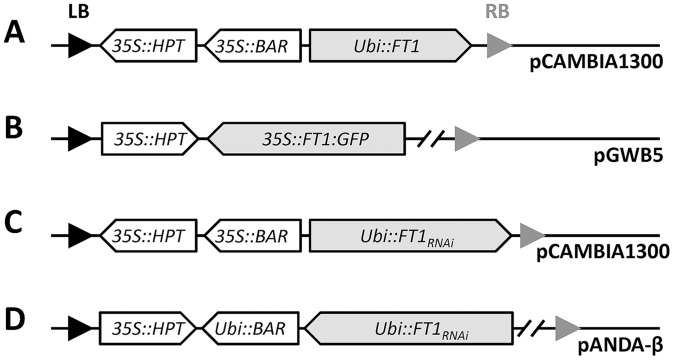
Figure 1. Schematic diagram of plasmids used in this study.
(A) Ubi::FT1 construct, (B) 35S::FT1:GFP construct. Overexpression constructs were developed in binary vectors pCAMBIA1300 and pGWB5. For Brachypodium FT1, the cDNA was cloned in both vectors. For wheat FT-B1, both the coding and genomic regions were cloned in pCAMBIA1300. (C and D) Ubi::FT1 RNAi constructs. (C) The Brachypodium FT1 RNAi trigger was cloned in the pCAMBIA1300-based vector. (D) The FT1 RNAi trigger from T. monococcum was cloned in the pANDA-based vector. In both constructs, expression of the selected RNAi trigger was driven by the maize Ubiquitin promoter (Ubi). The black and gray triangles indicate the left (LB) and right (RB) border repeats respectively.
For FT1 RNAi, we selected fragments from Brachypodium Bd21-3 cDNA (position 79 to 447) and G3116 cDNA (position 62 to 367). The selected RNAi triggers did not share any stretch of more than 18 identical nucleotides with other FT-like genes, preventing co-silencing of other FT-like genes. The Brachypodium FT1 RNAi trigger was cloned in the pCAMBIA1300-based vector (Figure 1C), and the wheat FT1 RNAi trigger was cloned in the pANDA-based vector (Figure 1D) [28]. In both constructs, expression of the selected RNAi triggers was driven by the maize Ubiquitin promoter (Ubi).
Generation of Transgenic Plants
Tissue culture and Agrobacterium-mediated transformation of Brachypodium was conducted as reported by Dr. Vogel and his colleagues [29]. Transgenic Brachypodium plants were established in soil in the growth chamber and in the greenhouse.
Protocols for the tissue culture and biolistic bombardment of wheat were adapted from a previous study [30]. Immature caryopses from T. aestivum cultivar Bobwhite were harvested two weeks after anthesis, sterilized with 70% (v/v) ethanol containing 0.05% (v/v) Tween 20 for 5 min, then with 20% (v/v) bleach supplemented with 0.05% (v/v) Tween 20 for 15 min, and washed 3–5 times using sterile distilled water. Immature embryos (ca. 1 mm long) were isolated from the sterilized caryopses, placed with the scutellum facing upward on the dissection media (MS base 4.3 g/L, maltose 40 g/L, thiamine-HCl 0.5 mg/L, L-asparagine 0.15 g/L, 2,4-D 2 mg/L, CuSO4 0.78 mg/L, Phytagel 2.5 g/L, pH 5.8), and maintained for 4–6 days at 22–23°C in the dark. Immature embryos were then treated for four hours on the high osmoticum media (MS base 4.3 g/L, maltose 40 g/L, sucrose 171.15 g/L, thiamine-HCl 0.5 mg/L, L-asparagine 0.15 g/L, 2,4-D 2 mg/L, CuSO4 0.78 mg/L, Phytagel 2.5 g/L, pH 5.8), and subjected to biolistic bombardment. Twenty hours after bombardment, immature embryos were transferred to recovery media (same as the dissection media), maintained for 2 weeks at 22–23°C in the dark. Embryo-derived calli were moved to the regeneration media (a dissection media supplemented with 0.1 mg/L 6-BA and 3 mg/L bialaphos) and maintained for two weeks in the growth chamber (22–23°C, 16 h light/8 h dark, light intensity of 25 μmol m−2 s−1). Regenerated shoots (2–3 cm) were transferred to the rooting media (a half-strength dissection media supplemented with 3 mg/L bialaphos), and maintained under the same environmental condition as for regeneration. Vigorous shoots with well-developed roots were established in soil in the greenhouse.
The biolistic bombardment was performed using the PDS-1000/He Particle Delivery System (Bio-Rad Laboratories, USA). To prepare three bombardments, 2.1 mg of microcarriers (Gold particles of 0.6 μm in diameter; Bio-Rad, USA) were measured into a 1.5 ml microcentrifuge tube, sterilized by mixing with 35 μl pure ethanol, recovered by spinning (12,000 rpm for 5 s) and removing the supernatant, rinsed in 200 μl ice-cold sterile distilled water, and collected by spinning and removing the supernatant. The pre-treated microcarriers were resuspended in 245 μl pre-chilled sterile water containing 20 μg plasmid DNA, and combined with another 250 μl pre-chilled CaCl2 (2.5 M). Where required, solutions in the previous steps were mixed thoroughly by pipetting. The microcarrier suspension was then supplied with 50 μl pre-chilled spermidine solution (1.45%, v/v) and mixed immediately by vortexing in the cold room (4°C) for 15–20 min. The plasmid-coated microcarriers were recovered by centrifugation (12,000 rpm for 10 s) and, following supernatant removal, were resuspended in 36 μl pure ethanol. For each bombardment, 10 μl gold suspension was loaded to the center of a macrocarrier disk (Bio-Rad), air-dried in the laminar flow hood, and placed in the microcarrier launch assembly under the 1100 psi rupture discs. Sixty immature embryos arranged in a 3.5-cm diameter circle were placed 6-cm below the macrocarrier assembly. The PDS-1000/He System was operated according to the manufacturer’s instruction. Bombardment conditions were 1,300 psi helium pressure and 25 mm Hg vacuum.
Putative transgenic plants were confirmed by PCR analysis. The presence of the selection marker (BAR or HPT) and other vector-specific fragments were tested in putative transgenic plants using PCR. Quantitative reverse transcription PCR (qRT-PCR) was used to quantify the downregulation of FT1 transcription in the RNAi transgenic plants and to confirm the expression of the FT1 RNAi constructs. In addition, transgenic wheat plants were confirmed by testing their resistance to 0.3% (v/v) Finale herbicide.
Mutant Screening and Genetic Analyses in Tetraploid Wheat
Genome-specific primers were designed for both FT-A1 and FT-B1 genes (Table 1) and were used to screen a tetraploid wheat TILLING population consisting of 1,384 EMS-treated individuals of the tetraploid wheat cultivar Kronos [31]. The screening was performed using a Cel I-based method described previously [31]. For the mutations described in this study, the first letter indicates the original base or amino acid and the last letter represents the mutant base or amino acid. The number in the middle indicates the position of the mutation counted from the ATG start codon in the genomic DNA sequence, or from the initial methionine in the predicted protein. For FT-A1, we selected mutant line T4-474 that carries a G655A mutation resulting in a premature stop codon. This mutant will be referred to hereafter as ft-A1. For FT-B1 no truncation mutations were found, so two different mutations resulting in amino acid substitutions at conserved amino acids were selected from lines T4-263 (C856T) and T4-344 (G98A), designated hereafter as ft-B1 263 and ft-B1 344, respectively. The predicted amino acid changes in each of the mutant lines are described in the results section.
Table 1. PCR primers used in the current study.
| Target | GenBank Acc. | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | Efficiency | Objective | References |
| TtFT-A1 | UCW_Tt_k51_contig_35084 1 | TCGATCTACACTAGGAAGAAGGAAG | GTGGGCCATGGGTAGG | ND | TILLING | Currentstudy |
| TtFT-B1 | UCW_Tt_k64_contig_13900 1 | GTGGGGCAACACTCATCATC | GGCTGGTGGCGTACGAG | ND | TILLING | Currentstudy |
| TtFT-A1 | UCW_Tt_k51_contig_35084 1 | AGACGTGCTGGACCCCTTT | GACTTGGAGCATCTGGGTCT | 98.50% | qRT-PCR | Currentstudy |
| TtFT-B1 | UCW_Tt_k64_contig_13900 1 | GGACGTGCTGGACCCCTTC | GACTTGGAGCATCTGGGTCT | 99% | qRT-PCR | Currentstudy |
| TaACTIN | UCW_Tt-k41_contig_5677 1 | ACCTTCAGTTGCCCAGCAAT | CAGAGTCGAGCACAATACCAGTTG | 98% | qRT-PCR | [34] |
| TaGI | AY6791154 | GAAGGTCAGAAGATGTGGAGAGTCAAC | GGCAGCGGATGGTAGGTGATAG | 95% | qRT-PCR | [24] |
| TaFT1 | CD881060 | GCCGGTCGATCTATACTA | TCCTGTTCCCGAAGGTCA | 101% | qRT-PCR | [24] |
| TaFT2 | BT009051 | TTTCTACACGCTGGTGATGG | GTGACCAGCCAGTGCAAGTA | 96% | qRT-PCR | [21] |
| TaFT3 | IWGSC_1AL_913428 2 | GTACTTGCACTGGATGGTGTC | CATCTGGTGCAAAAACTGT | 93% | qRT-PCR | Currentstudy |
| TaFT4 | IWGSC_2AS_5252557 2 | TGGATCCTGATGCGCCTAA | CAGTCACCATCCAGTGCAGGTA | 109% | qRT-PCR | Currentstudy |
| TaFT5 | IWGSC_5AL_2803506 2 | ACGGTTTTTGCACCGGACA | GGCAGCGGCAATGTTGAG | 100% | qRT-PCR | Currentstudy |
| TaFT6 | IWGSC_6AS_4388307 2 | GATATGCATGGCGGTTTCTC | CCAAGGAGTCGTTCGACATT | 93% | qRT-PCR | Currentstudy |
| TaVRN1 | JF965395 | AAGAAGGAGAGGTCACTGCAGG | GGCTGCACTGCCGCA | 99% | qRT-PCR | [7] |
| TmACTIN | AF326781 | GCCATGTACGTCGCAATTCA | AGTCGAGAACGATACCAGTAGTACGA | 99% | qRT-PCR | [33] |
| BdGI | Bradi2g05226 | TACGGATGGGATGCTTGTTG | CGGCACTTCAGCAGATTCG | 99% | qRT-PCR | Currentstudy |
| BdFT1 | Bradi1g48830 | CACACTACACACACGCAAGTACTGT | CAGCACGTCCCCCACAA | 99% | qRT-PCR | Currentstudy |
| BdFT2 | Bradi2g07070 | TGGTTGTGATGGTCCGTTTG | AGACAGAACCGACTTGCTAGAAATTAC | 98% | qRT-PCR | Currentstudy |
| BdFT3 | Bradi2g49795 | CCCTGGGACAACTGGAGCTA | TTCTTGGTTCTGGTCTTTCGTAGA | 105% | qRT-PCR | Currentstudy |
| BdFT4 | Bradi1g38150 | TGGGCGGGAGATCGTAAC | CGGTGGATGCCCATGGT | 106% | qRT-PCR | Currentstudy |
| BdFT5 | Bradi2g19670 | GAAGGTGGATCGGGTGGAA | GCTGTCTAGTCTTTACTCCCCTTGA | 103% | qRT-PCR | Currentstudy |
| BdFT6 | Bradi3g08890 | GCGAGGACCTCAGCGTAACA | GCCGGGCTCTCGTAGCA | ND3 | qRT-PCR | Currentstudy |
| BdVRN1 | GT846767 | GTCGCGCTCATCATCTTCTC | TGCATAGGAGTAGCGCTCATAG | 100% | qRT-PCR | [23] |
| BdACTIN | Bradi4g41850 | CCTGAAGTCCTTTTCCAGC | AGGGCAGTGATCTCCTTGC | 99% | qRT-PCR | [53] |
Sequences from tetraploid wheat Kronos [54];
Sequences from International Wheat Genome Sequencing Consortium (IWGSC) (Ensembl; http://plants.ensembl.org/index.html);
ND: Not Determined.
We backcrossed ft-A1 three times to wild type Kronos and then crossed it with an M3 plant of the mutant line ft-B1 263 to generate a BC1F2 population segregating for both mutations. Epistatic interactions between FT-A1 and FT-B1 were analyzed in this population using a factorial ANOVA. Data from this population was also used to calculate the degree of dominance for FT-A1 and FT-B1 using the formula: D = (2X2−X1−X3)/(X1−X3) [32] where X1, X2 and X3 are the heading time values, respectively, of the plants homozygous for the mutant late flowering allele (ft), the heterozygotes, and the plants homozygous for the wild type early flowering allele (FT). The degree of dominance for FT-A1 was calculated using only ft-B1 263 homozygous plants, and the degree of dominance for FT-B1 was calculated using only ft-A1 homozygous plants.
Mutant line ft-B1 344 was backcrossed to Kronos and a separate BC1F2 segregating population was developed to evaluate the effect of this mutation on heading time. Both segregating populations were evaluated simultaneously for heading time in the same greenhouse experiment.
The transcript levels of FT-A1 and FT-B1 homoeologs were compared in three-week-old wild type Kronos plants grown in growth chambers under LD (16 h light, light intensity of 90 μmol m−2 s−1) at 16–20°C.
Quantitative Reverse Transcription PCR (qRT-PCR) Analysis
Gene expression in FT1 RNAi transgenic plants was determined from six biological replicates. The youngest leaves of adult plants were collected at 12∶00 p.m. for Brachypodium and 4∶00 p.m. for hexaploid wheat. Total RNA was extracted from leaf tissues using the Trizol method (Life Technologies, Grand Island, NY, USA) according to the manufacturer’s instructions. cDNA templates were prepared using the Fermentas First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). In total, eight target genes were studied (Table 1). Ten-fold serial dilutions of cDNA templates were used to test the amplification efficiency of each primer pair. In Brachypodium and in Bobwhite transgenic wheat and control lines, qRT-PCR was performed using the FastStart SYBR Green Master (Roche Applied Science, Indianapolis, IN, USA) on the StepOnePlus Real-Time PCR Systems (Life Technologies). The amplification conditions were one cycle of 10 min at 95°C, 40 cycles of two consecutive steps of 15 s at 95°C and 1 min at 60°C and a standard dissociation protocol. ACTIN was used as an endogenous control using primers designed by Fu, et al. (Table 1; [33]).
In tetraploid wheat and in FT1 HOPE lines, the youngest leaf was collected at 10∶00 a.m. and RNA was extracted using the Spectrum Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO, USA). cDNA was synthesized using the High Capacity cDNA Reverse Transcription kit (Life Technologies) and used in qRT-PCR reactions performed on a 7500 Fast Real-Time PCR System (Life Technologies). Transcript levels of FT-A1 and FT-B1 homoeologs in tetraploid wheat were compared using genome specific primers (Table 1). ACTIN was used as an internal control using primers originally designed by Uauy, et al. (Table 1; [34]). The qRT-PCR conditions were one cycle of 20 s at 95°C and 40 cycles of two consecutive steps of 3 s at 95°C and 30 s at 60°C.
Transcript levels of target genes were calculated using the formula 1000*2(ACTIN CT – TARGET CT), which indicates the relative number of target molecules per 1000 molecules of ACTIN. The expression data was analyzed using SAS version 9.0 (SAS Institute Inc, Cary, NC, USA). Data that did not meet the assumptions of the ANOVA was transformed using power transformations to restore homogeneity of variances (Levene’s test) and normality of residuals (Shapiro-Wilk test). Independent transgenic events were compared with the wild type control using Dunnett’s tests, and the information was summarized using a contrast comparing the wild type versus all transgenic lines. Graphs were prepared using the GraphPad Prism version 5.01 (GraphPad Software, San Diego, CA, USA).
Pollen Staining
The viability of pollen grains in FT1 transgenic plants was evaluated using a simplified staining protocol [35]. Mature but non-dehiscent anthers were collected and submerged in 100 μl staining solution for 24 h at room temperature and in darkness. After rinsing in distilled water, anthers were transferred to a fresh slide containing a drop of distilled water, and then inspected under the stereomicroscope. To measure the percentage of viable pollen, anthers were gently pressed to release pollen grains. Pollen grains stained magenta-red were viable, and those stained blue-green were non-viable. Two-hundred pollen grains within the microscopic view, performed in triplicate, were used to calculate the proportion of viable pollen.
Results
Overexpression of FT1 Promoted Floral Organogenesis During Tissue Culture
To study the effect of increased FT1 expression on floral development, we developed transgenic Brachypodium and wheat plants overexpressing FT1. In the Brachypodium transformation experiment using the FT1 OE construct for Bd21-3 FT1cDNA driven by the maize UBIQUITIN promoter (Figure 1A), none of the 180 calli regenerated under LD (16 h light/8 h dark) and only two out of 100 calli regenerated under SD (8 h light/16 h dark), grew weakly, and eventually died. In the second Brachypodium transformation experiment using the fusion construct of Bd21-3 FT1 and GFP driven by the 35S promoter (Figure 1B), six of 280 infected calli showed regenerated shoots that immediately developed floral organs under LD (Figure 2A–C). As a result of the limited development of vegetative tissue, none of the T0 plants produced seeds, and they did not survive when transplanted to soil. We then adjusted the light conditions during callus differentiation and shoot regeneration to a SD photoperiod and weak light intensity (29 μmol m−2 s−1). Under these conditions, the floral organogenesis was delayed and there was an increase in vegetative growth, but it was still insufficient to support the production of seeds. The lack of normal vegetative tissue also precluded the use of the GFP tag to study the localization of the FT protein in Brachypodium leaves.
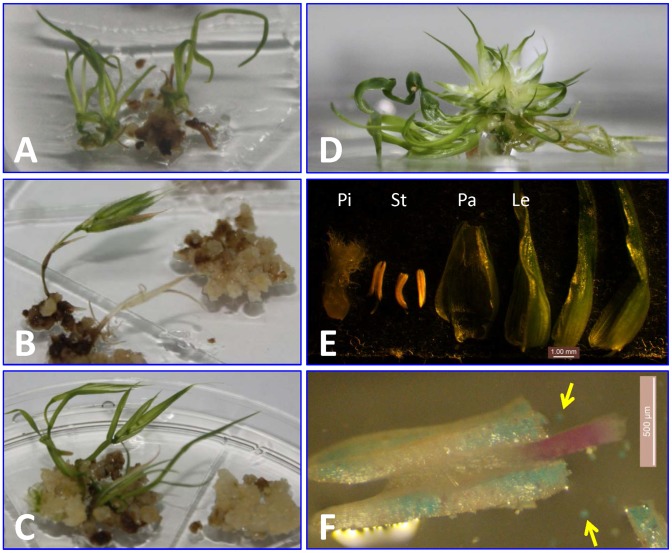
Figure 2. FT1 overexpression promotes floral organogenesis.
(A–C) Brachypodium 35S::BdFT1:GFP, (A) Shoot regeneration of non-transgenic control calli (B) direct spike formation from transformed calli and (C) rudimentary leaves associated with spikelet formation from transformed calli. (D–F) Wheat Ubi::cFT-B1. (D) Cluster of florets surrounded by rudimentary leaves in a transformed callus. (E) Different floral organs: lemma (Le), palea (Pa), pistil (Pi) and stamen (St). The additional organs seem to be glumes but it was difficult to determine because of the close clustering of multiple florets. (F) Anther with regions of non-viable pollen (blue color after pollen staining).
In common wheat, we tested two different FT1 OE constructs, both driven by the maize Ubiquitin promoter: the first one using the FT-B1 coding region (Ubi::cFT-B1) and the second one using the FT-B1 genomic region (Ubi::gFT-B1) from tetraploid wheat Langdon (Figure 1A). Of the 1,876 calli bombarded with the Ubi::cFT-B1 construct, 12 developed floral organs during culture. Of the 523 calli bombarded with the Ubi::gFT-B1 construct, three flowered during the culture stage. PCR analysis confirmed that all early-flowering T0 plants were positive for the presence of the transgene (data not shown). In both cases, calli were maintained in recovery culture under dark conditions for four weeks and were then transferred to regeneration media under LD. Green shoots appeared five days later, and clusters of florets developed 3 to 4 weeks later (Figure 2D). At this stage, florets were opened wide and exhibited a normal feathery stigma but with smaller and shriveled stamens. Most florets had complete structures (including lemma, palea, pistil and stamens, Figure 2E), but some florets did not develop stamens. In comparison to wild type plants, the anthers of the transgenic plants had fewer pollen grains and a large proportion of non-viable pollen, as determined by pollen staining (Figure 2F). Similar to the transgenic Brachypodium plants, there was limited leaf development and none of the T0 plants produced seeds in culture or were able to survive when transplanted to soil.
Downregulation of FT1 Delayed Heading Time of Transgenic Plants
To determine the effect of reduced FT1 expression on flowering development, we developed Brachypodium and wheat FT1 RNAi constructs that target only the FT1 gene. Of the 280 Brachypodium calli transformed with the Brachypodium FT1 RNAi construct (Figure 1C), a total of 38 calli generated putative transgenic plants. PCR analyses demonstrated that all putative transgenic plants were positive for the FT1 RNAi construct. All Brachypodium transgenic T0 plants grew vigorously, but failed to flower (Figure 3A) even after two weeks of vernalization at 4°C, which is sufficient to induce flowering in the non-transgenic Bd21-3 control plants.
Figure 3. Silencing of FT1 by RNAi delays heading time.
(A) Heading was prevented in transgenic FT1 RNAi Brachypodium and (B) delayed in FT1 RNAi transgenic wheat. (C) Wheat transgenic plants at booting stage. (D) Some spikes had difficulty in emerging from the leaf sheath (leaf sheath opened manually in this picture). (E) Complete floral organs from transgenic wheat flowers (stigmas failed to open in some transgenic plants).
In the common wheat cultivar Bobwhite, the FT1 RNAi construct shares a continuous stretch of at least 179 identical nucleotides with the FT-A1, FT-B1, and FT-D1 homoeologs, and therefore, is expected to silence all three copies simultaneously. Of the 1,675 wheat calli bombarded with the FT1 RNAi construct, 13 independent plants were confirmed to be transgenic from their resistance to the herbicide Finale and by PCR analysis. Transgenic lines showed heterogeneity in the effect of the transgene on heading time, floral organ morphology and fertility (Figure 3B–D, Table 2). Ten of the 13 transgenic plants showed moderate delays in heading time ranging from 2 to 4 weeks, two showed very late heading time (lines ‘1965’ and ‘2547’), and one (line ‘2548’) remained vegetative for ten months, before dying without flowering. Spikelets in line ‘2547’ had two complete florets, but additional florets were undeveloped. Of the two intact florets, each consisted of one pistil and three stamens, but the stigmas remained closed (Figure 3E) and the anthers stayed green and did not shed pollen. Of the two late transgenic lines only ‘1965’ set seeds.
Table 2. Heading date and floral characteristics of wheat FT1 RNAi transgenic plants.
| Plant ID | Generation | Heading Date | Spikelets | Stigma | Anthers | Pollen Viability | Seed Setting |
| Bobwhite | WT | Normal | Normal | Bifid feathery | Normal | 90–98% | Normal |
| Ten T0 linesa | T0 | +14 to 28 d | Normal | – | – | – | Yes |
| 1965 | T0 | +4 m | Normal | Variabled | Small | – | Yes |
| 2547 | T0 | +6 m | Variablec | Closed | Small | – | None |
| 2548 | T0 | Stay green | – | – | – | – | None |
| Three T1 linesb | T1 | Normal | Normal | Normal | Normal | – | Yes |
| 140D-1 | T1 | +14 d | Normal | Variabled | Small | 47–66% | Yes |
| 152E-1 | T1 | +14 d | Normal | Variabled | Small | 43–78% | Yes |
| 152E-2 | T1 | +17 d | Normal | Variabled | Small | 11–32% | Yes |
| 1965–2 | T1 | +10 d | Normal | Variabled | Small | 45–67% | Yes |
| 1965–5 | T1 | +20 d | Variablec | Variabled | Small | 79–90% | None |
The ten T0 lines include ‘9′, ‘78A’, ‘116’, ‘140D’, ‘152E’, ‘157B’, ‘1381’, ‘1382’, ‘1384’ and ‘1828’;
The three T1 lines include ‘157B’, ‘1381’ and ‘1382’;
The first and second florets are normal, others deficient;
Stigmas are bifid and feathery but open less widely than in the wild type.
Progeny tests including 5 to 10 T1 plants were performed for six independent transgenic lines, including the late-flowering line ‘1965’ and five moderately late-flowering lines. The T1 progeny of the late-flowering T0 transgenic plant ‘1965’ were not as late as the original T0 plant (only 10–20 days delay compared to the wild type) suggesting that late flowering of the T0 plant was not determined solely by the downregulation of FT1. Unfortunately, no seeds were available for the T0 transgenic lines showing the greatest delay in flowering (lines ‘2547’ and ‘2548’) and, therefore, we were unable to verify the linkage between these late flowering phenotypes and the transgene.
Progeny for lines ‘140D’, ‘152E’ and ‘1965’ segregated for early and late heading time (14 to 20 days later than the Bobwhite control). Progeny of ‘152E’ developed relatively normal florets and set seeds. In a few lines, e.g. ‘140D-1′ spikes failed to emerge from the sheath of the flag leaf resulting in abnormal curling of the spike and awns (Figure 3D). Additionally, the florets developed pistil and stamen, but some florets had abnormal stigmas which failed to open after heading and did not set seeds. All plants from the progeny of lines ‘157B’, ‘1381’ and ‘1382’ headed at the same time as the wild type control plants suggesting that they were not functional transgenic lines (Table 2). In summary, FT1 RNAi transgenic wheat lines flowered 2 to 4 weeks later than the wild type control, exhibited reduced pollen viability and in some cases mature stigmas did not open as widely as those in control plants (Table 2).
Mutations in FT-A1 and FT-B1 Delay Flowering in Tetraploid Wheat
We screened a TILLING population of the tetraploid wheat cultivar Kronos and identified 38 mutant alleles for FT-A1 and 13 for FT-B1, and selected three for functional characterization (Table 3). The G655A mutation present in the selected ft-A1 mutant disrupts the splice site located at the beginning of the second intron. Sequencing of the resulting ft-A1 cDNA using homoeolog-specific primers confirmed the elimination of the splice site, which resulted in a 4-bp insertion and an in-frame premature stop codon at position 88 (W88*). This premature stop codon eliminates the last 90 amino acids (50.8% of the protein), and almost certainly produces a non-functional protein.
Table 3. Summary of the ft1 mutants in tetraploid wheat cultivar Kronos.
| Line | Gene | Nt Mutationa | Pr Mutation b | BLOSUM 62c | At/Os/Zm d | Hv/Ta/Tm/Tt d | Hv/Tt d | ||||
| FT1 | FT1 | FT2 | FT3 | FT4 | FT5 | FT6 | |||||
| T4-474 | TtFT-A1 | G655A | W88* | N/A | W | W | W | W | W | W | W |
| T4-263 | TtFT-B1 | C856T | P77S | −1 | P | P | P | P | P | P | P |
| T4-344 | TtFT-B1 | G98A | G33E | −2 | G | G | A | N | D | N | D |
Mutations in the DNA (Nt, position is relative to the start codon ATG);
Mutations in the predicted protein (Pr) (counted from the initial methionine);
BLOSUM 62 scores [36];
Amino acid present in other species: Arabidopsis (At), Hordeum vulgare (Hv), Oryza sativa (Os), Triticum aestivum (Ta), T. monococcum (Tm), T. turgidum (Tt), and Zea mays (Zm). All homoeologous alleles were included in the comparison for the polyploidy wheat species.
No truncations or splice site mutations were identified for FT-B1, so two substitution mutations within the PEBP motif were selected based on high evolutionary conservation of the targeted amino acid and negative BLOSUM62 scores, which are predictive of altered structural and functional properties [36]. In mutant line ft-B1 263, a C856T mutation resulted in a change between proline and serine (P77S, BLOSUM62 score = −1). The proline at position 77 is highly conserved in FT1 orthologs in Arabidopsis, maize, rice, diploid, tetraploid and hexaploid wheat and in all six FT-like proteins (FT1 to FT6) in barley and wheat (Table 3). Mutant line ft-B1 344 carries a G to A mutation at position 98 (G98A), which results in the amino acid change G33E (BLOSUM62 score = −2). The original amino acid (glycine) is conserved in the FT1 orthologs in Arabidopsis, maize, rice, barley, diploid, tetraploid and hexaploid wheat. In the other FT-like proteins this amino acid position is substituted by alanine, asparagine or aspartic acid, but in no case by glutamic acid as in the G33E mutation (Table 3).
To quantify the effect of the ft-A1 truncation mutation, the ft-B1 263 substitution mutation, and their interaction we genotyped 110 plants from a BC1F2 population segregating for these two mutations and determined their heading time under LD without vernalization (Kronos has a spring growth habit). The wild type alleles for early flowering showed partial dominance over the mutant alleles, with an estimated degree of dominance of approximately 0.46 and 0.59 for FT-A1 and FT-B1, respectively.
Based on this partial dominance we decided to merge the homozygous wild type and heterozygous classes into a single non-mutant class. This strategy has the advantage of simplifying the analysis and description of the interactions between these two genes, and of increasing the power of the statistical analyses. The resulting 2×2 factorial ANOVA (using FT-A1 and FT-B1 as factors) revealed significant differences in heading time both for FT-A1 (P = 0.0075) and FT-B1 (P<0.0001), as well as a significant interaction (P = 0.0143, Figure 4A) between the two genes. This interaction indicates that the effect of each FT1 homoeolog on flowering time is dependent on the allele present in the other homoeolog.
Figure 4. ft1 TILLING mutants.
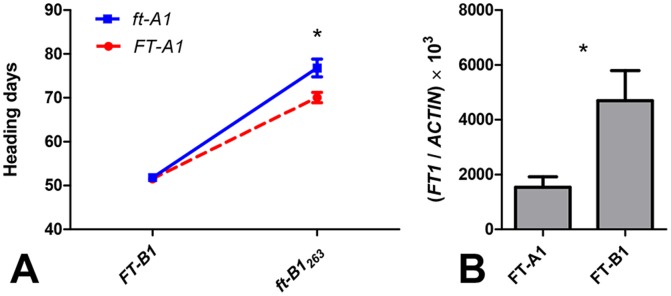
(A) Interaction plot showing the effect of ft-A1 and ft-B1 263 mutations on heading time in a BC1F2 population segregating for both genes. Differences in heading time between the FT-A1 alleles were significant only for the homozygous ft-B1 263 mutant plants. (B) Transcript levels of FT-A1 and FT-B1 homoeologs in three-week-old wild type Kronos. The expression level of FT-B1 was significantly higher than that of the FT-A1 (P = 0.017). ACTIN was used as an internal control. Samples were harvested at 10∶00 a.m. Asterisks indicate P values of Student’s t-tests: * = P<0.05.
To describe these interactions, we analyzed the simple effects of each FT1 homoeolog within the two classes of the other FT1 homoeolog. The FT-B1 alleles showed highly significant differences in heading time (P<0.0001) within both alleles of FT-A1, but the differences between the adjusted means of the FT-B1 alleles were 34% larger in the lines homozygous for the mutant ft-A1 allele (25.0 day delay in flowering) than in the lines homozygous or heterozygous for the wild type FT-A1 allele (18.6 day delay). The effects of the FT-A1 alleles on heading time were smaller than those of the FT-B1 alleles, and the differences were significant only within the homozygous ft-B1 263 mutant class (6.7 day delay; P = 0.0374). In all cases, plants carrying mutant ft1 alleles flowered later than plants homozygous or heterozygous for the wild type alleles. Plants homozygous for both ft-A1 and ft-B1 263 mutant alleles flowered on average 25.0 days later than plants homozygous or heterozygous for the two wild type alleles, confirming an important role of FT1 in the acceleration of wheat flowering under LD.
To characterize the effect of the ft-B1 344 mutation (G98A) on heading time we genotyped and phenotyped 23 BC1F2 plants segregating for the ft-B1 344 mutation in a genetic background fixed for the wild type FT-A1 allele. Plants homozygous for the wild type FT-B1 allele headed on average in 43.5 days while plants homozygous for the mutant ft-B1 344 allele headed in 51.2 days (7.7 day delay). This difference was smaller than that detected between homozygous lines of wild type and ft-B1 263 mutation (20.4 day delay within the same homozygous wild type FT-A1 background), suggesting that the ft-B1 344 mutation has a less deleterious effect on FT-B1 function than the ft-B1 263 mutation.
The predicted proteins encoded by the FT-A1 and FT-B1 genes in tetraploid wheat are identical, so we hypothesized that the larger effect of the ft-B1 263 mutant relative to the ft-A1 truncation mutant on heading time could be associated with relative differences in their respective expression levels. To test this hypothesis, we designed genome-specific primers for FT-A1 and FT-B1 and quantified their transcript levels using qRT-PCR (Table 1) in three-week-old wild type Kronos plants. Transcript levels of the FT-B1 homoeolog were three-fold higher than those of the FT-A1 homoeolog (Figure 4B), which correlates with the stronger effect of the FT-B1 locus on heading time.
Expression of FT-like Genes in Transgenic FT1 Plants
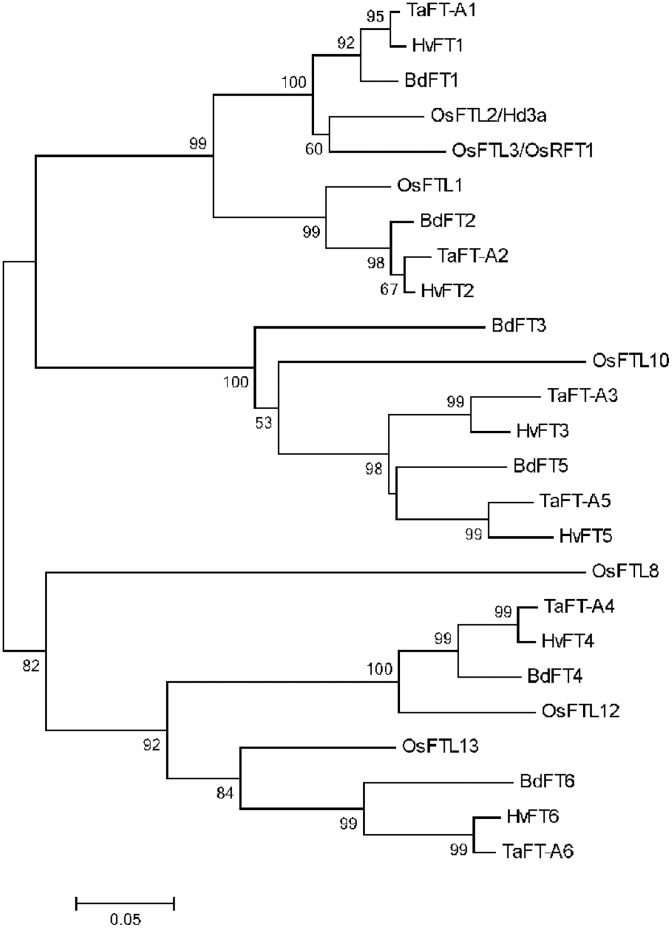
Based on the observation that the transcript levels of FT2 were altered in transgenic wheat plants with higher transcript levels of FT1 [21] we decided to investigate the effect of FT1 down- and up-regulation on the five closest paralogs to FT1. To provide an evolutionary framework for the different wheat and Brachypodium FT-like genes included in the expression analyses, we first characterized their phylogenetic relationships (Figure 5). There are multiple FT-like genes in both Brachypodium and wheat [12], but only the closest five are included in this study. The predicted proteins from Brachypodium, wheat, barley and rice were grouped into five clusters. Both the FT1 and FT2 clusters include homologs from all four species, which suggests an ancient duplication that predates the divergence between rice and the temperate grasses. The FT3 and FT5 clusters are more closely related to each other than to the FT4 cluster, which includes two closely related proteins, FT4 and a previously undescribed protein, which we have named FT6 (Table 1). Barley and Brachypodium homologs of this novel FT protein were also identified (Figure 5).
Figure 5. Phylogenetic analysis of FT-like genes in temperate grasses.
Phylogenetic analysis was performed using the full-length proteins. A neighbor-joining tree was constructed using pairwise deletions and 1,000 bootstrap iterations with the program MEGA 5.0 [52]. The scale bar 0.05 represents 5% base substitution. Bootstrap numbers larger than 50 are shown in the respective nodes. To simplify the tree, only the wheat A-genome homoeologs of wheat were included. Accession information: BdFT1 (Bradi1g48830), BdFT2 (Bradi2g07070), BdFT3 (Bradi2g49795), BdFT4 (Bradi1g38150), BdFT5 (Bradi2g19670), BdFT6 (Bradi3g08890), HvFT1 (DQ100327), HvFT2 (DQ297407), HvFT3 (DQ411319), HvFT4 (DQ411320), HvFT5 (EF012202), HvFT6 (morex_contig_54196), OsFTL1 (Os01g11940), OsFTL2/Hd3a (Os06g06320), OsFTL3/RFT1 (Os06g06300), OsFTL8 (Os01g10590), OsFTL10 (Os05g44180), OsFTL12 (Os06g35940), OsFTL13 (Os02g13830), TaFT-A1 (CD881060), TaFT-A2 (BT009051), TaFT-A3 (IWGSC_1AL_913428), TaFT-A4 (IWGSC_2AS_5252557), TaFT-A5 (IWGSC_5AL_2803506), TaFT-A6 (IWGSC_6AS_4388307). HvFT6 sequence is from the International Barley Sequencing Consortium (IBSC, http://webblast.ipk-gatersleben.de/barley/).
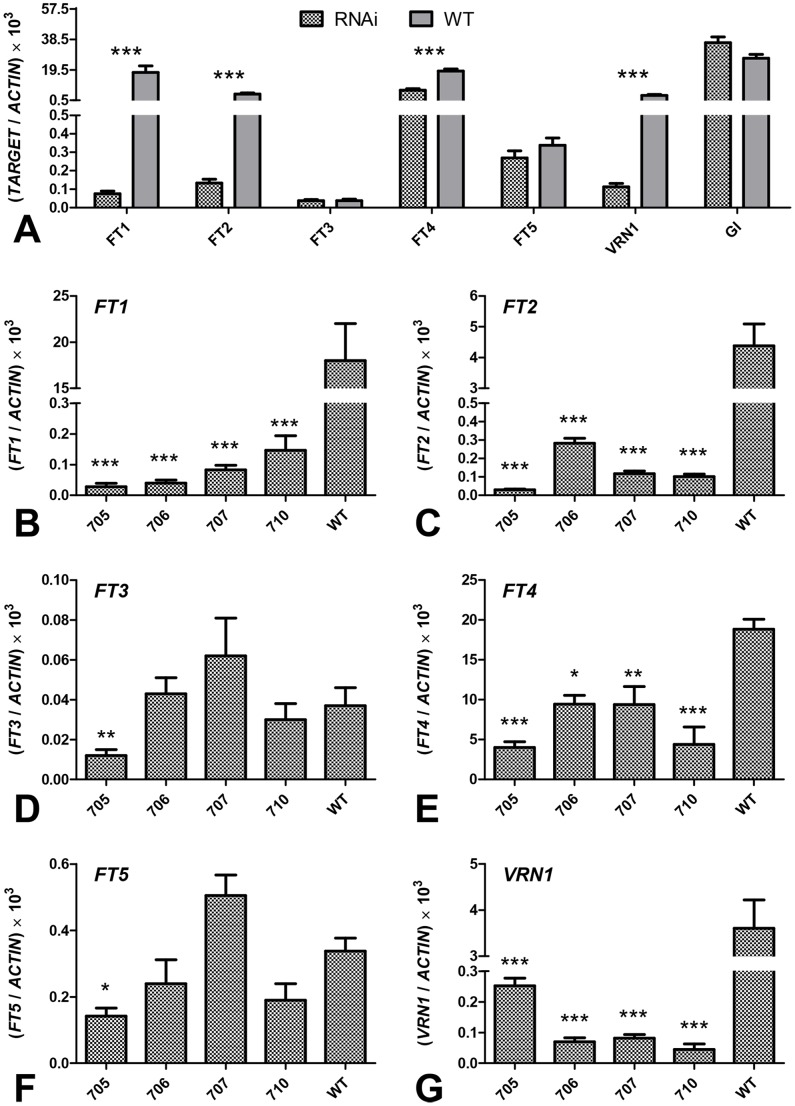
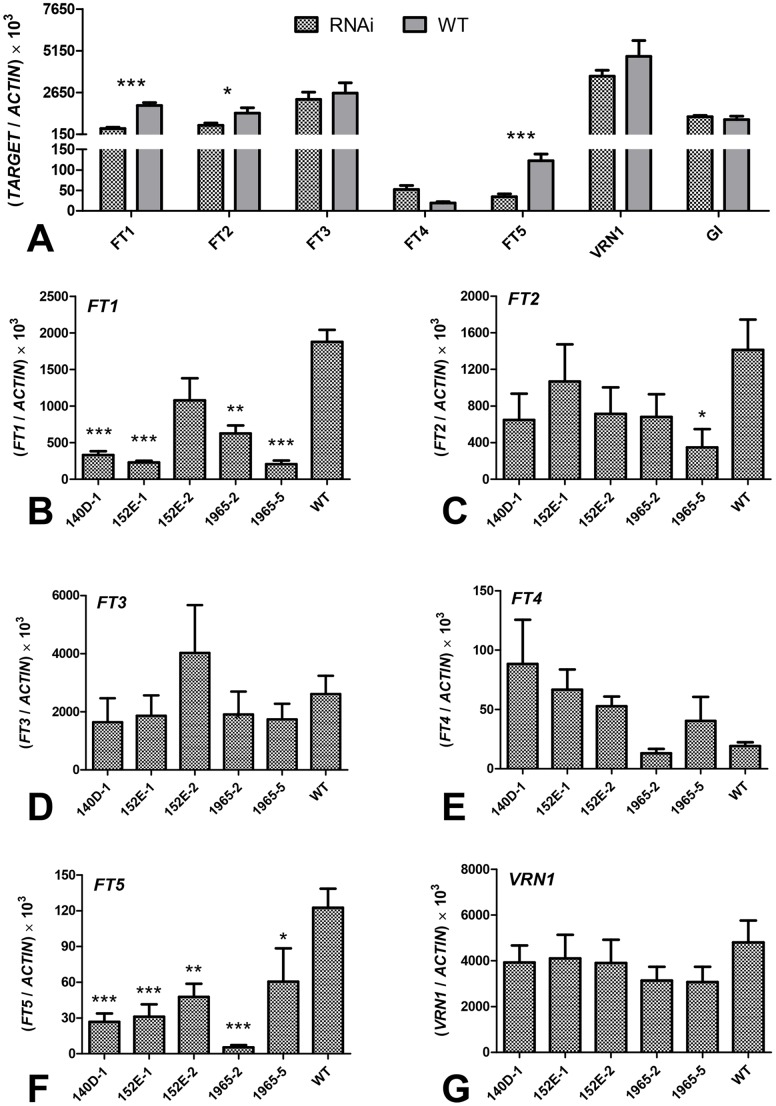
Brachypodium FT1 RNAi T0 plants ‘705’, ‘706’, ‘707’, and ‘710’ grown under LD showed a significant reduction in FT1 transcript levels (<1%, P<0.001) relative to the wild type control (Figure 6A–B). The same transgenic plants also showed a significant downregulation of the FT-like genes FT2 (average 3% of wild type, P<0.001, Figure 6C) and FT4 (average 36% of wild type, P<0.001, Figure 6E), but their expression of FT3 or FT5 was not significantly different from the wild type (Figures 6A,D,F). The expression of FT6 was nearly undetectable in both wild type and transgenic plants (data not shown). The FT1 RNAi transgenic plants also showed a strong downregulation of the FT1 downstream target VRN1 (∼3% of wild type, P<0.001, Figure 6G), which may explain the non-flowering phenotype of these lines. As expected, no significant effects were detected in the expression of GIGANTEA (GI) (Figure 6A), an upstream regulator of FT in the photoperiodic pathway in Arabidopsis [37].
Figure 6. Transcript levels of target genes in T0 Brachypodium FT1 RNAi lines.
(A) Comparison between the average of four transgenic plants (RNAi) and the wild type control using contrasts. (B–G) Comparison between individual transgenic lines and wild type using Dunnett’s test. (B) FT1, (C) FT2, (D) FT3, (E) FT4, (F) FT5, (G) VRN1 (gene regulated by FT1). ACTIN was used as the internal control. Samples were harvested at 12∶00 p.m., approximately one week after wild type control plants began to flower. Asterisks indicate P values: * = P<0.05; ** = P<0.01; *** = P<0.001.
In the five selected wheat FT1 RNAi T1 transgenic lines (‘140D-1′, ‘152E-1′, ‘152E-2′, ‘1965-2′, and ‘1965-5′), FT1 transcript levels were significantly reduced relative to the levels observed in the wild type Bobwhite (∼26%, P<0.001, Figure 7A–B). The downregulation of FT1 was associated with significant reductions in the transcript levels of FT2 (49%, P<0.05, Figures 7A,C) and FT5 (28%, P<0.001, Figures 7A,F). No significant differences between mutant and wild type lines were detected for FT-like genes FT3 and FT4 (Figures 7A,D–E) or for the GI upstream control (Figure 7A). As in Brachypodium, the expression of the wheat FT6 gene was also nearly undetectable in both wild type and transgenic plants (data not shown). Transcript levels of VRN1 were slightly reduced (76% of wild type) in the transgenic plants, but these differences were not significant (Figures 7A,G). The smaller differences in VRN1 transcript levels and in flowering time between the wheat FT1 RNAi and wild type compared with those observed in Brachypodium, might be associated with the presence of dominant VRN1 alleles for spring growth habit in both the Bobwhite transgenic and control plants (Vrn-A1, Vrn-B1 and Vrn-D1; [38]).
Figure 7. Transcript levels of target genes in transgenic T1 wheat RNAi plants and the non-transgenic wheat control.
(A) Comparison between the average of five transgenic plants (RNAi) and the wild type control using contrasts. (B–G) Comparison between individual transgenic lines and wild type using Dunnett’s test. (B) FT1, (C) FT2, (D) FT3, (E) FT4, (F) FT5, (G) VRN1 (gene regulated by FT1). ACTIN was used as the internal control. Samples were harvested at 4∶00 p.m., when the wild type controls began to flower. Asterisks indicate P values: * = P<0.05, ** = P<0.01, *** = P<0.001.
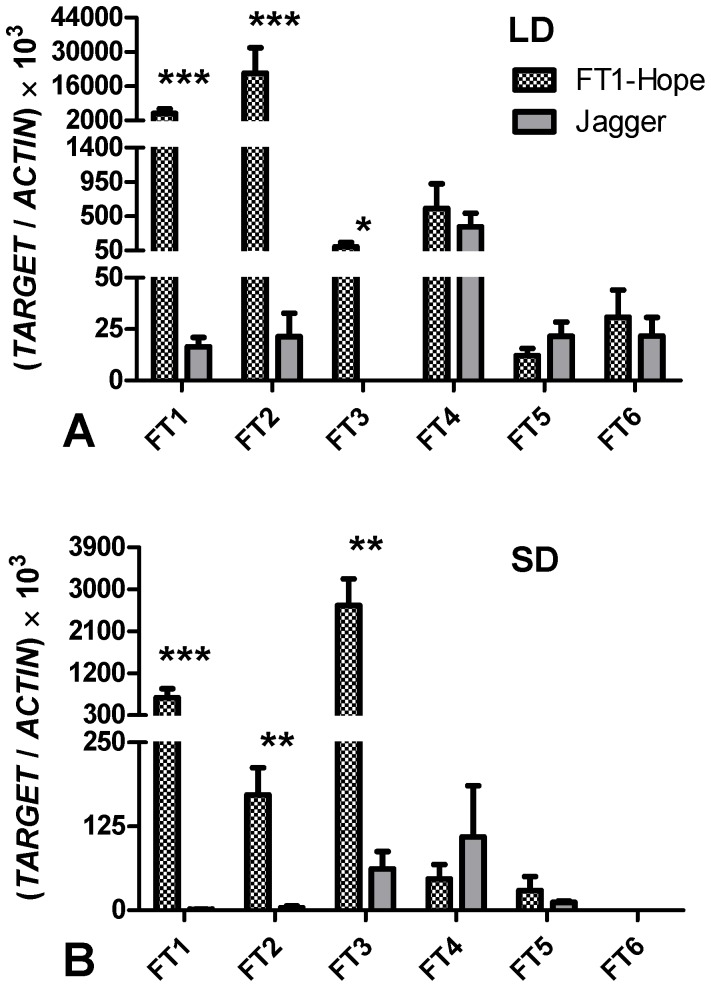
To further test the effect of FT1 (or of the developmental changes induced by FT1) on the expression of other FT-like genes, we quantified the transcript levels of these genes in wheat transgenic plants carrying the highly-expressed Hope FT-B1 allele [7]. In the transgenic FT1 HOPE lines, the significant increase in FT1 transcript levels was associated with a significant upregulation of FT2 and FT3 both under LD (Figure 8A) and SD (Figure 8B) photoperiods. Expression levels of FT4, FT5 and FT6 were not significantly different between the wild type and FT1 HOPE lines. Taken together these results indicate that the developmental changes induced by FT1 are associated with changes in the expression of other FT-like genes.
Figure 8. Expression of FT-like genes in FT1 HOPE transgenic wheat.
The experiment was performed on transgenic lines (FT1 HOPE) and the non-transgenic control Jagger. Leaf tissues were collected from two independent experiments, including plants grown under a (A) LD photoperiod for five weeks and (B) plants grown under a SD photoperiod for six weeks. ACTIN was used as the internal control. Samples were harvested at 10∶00 a.m. Asterisks indicate P values: * = P<0.05, ** = P<0.01, *** = P<0.001.
Discussion
FT1 is a Strong Promoter of Flowering Initiation in Both Brachypodium and Wheat
In the temperate grasses, the shoot apical meristem is located at the base of the plant covered by multiple layers of nested leaf primordia, sometimes below the soil and/or a layer of dead leaves, and is thus relatively isolated from some environmental signals. In contrast, leaves are broadly exposed and act as the main sensory organ for monitoring changes in the environment. FT1 and possibly other FT-like genes play a central role in the integration and transport of these signals [7], [18]. The convergence of multiple signaling pathways in the regulation of these genes is likely related to the distinctive ability of their encoded proteins to move through the phloem transporting environmental signals from the leaves to the shoot apical meristems [17], [20].
In this study we show that the ectopic expression of FT1 under constitutive promoters results in precocious flowering of differentiating wheat and Brachypodium calli, bypassing the generation of normal vegetative tissues. Similar results have been reported before for both Brachypodium and wheat. Two recent studies have shown that overexpression of FT1 (and also of FT2) in Brachypodium results in precocious flowering [25], [26]. Similarly, transformation of the FT-D1 wheat homoeolog driven by the 35S promoter into the spring wheat variety ‘Norin 61′ [24], or of a highly expressed FT-B1 allele driven by its native promoter into the winter wheat Jagger (FT1 HOPE) [7], accelerate wheat heading time. Taken together, these results demonstrate that FT1 is a strong activator of the flowering pathway in both wheat and Brachypodium.
FT1 has the ability to induce the transcription of VRN1 meristem identity gene even in the absence of vernalization. In varieties with a winter growth habit, chromatin in VRN1 regulatory regions is in a repressed state before vernalization and cold treatment promotes a more active state leading to the upregulation of VRN1 [39]. We show here that the overexpression of FT1 in the vernalization-responsive Brachypodium accession Bd21-3 [26], [40] results in extremely early flowering, even in the absence of vernalization. Similarly, the introgression of natural FT1 alleles expressed at high levels in early developmental stages for both wheat [7] and barley [41] into varieties with a winter growth habit also results in early flowering in the absence of vernalization.
Downregulation of FT1 Showed Stronger Effects in Brachypodium than in Wheat
Results from the FT1 overexpression experiments presented here and from previous studies [7], [24], [25] indicate that the expression of FT1 is sufficient to induce flowering in both Brachypodium and wheat. However, these studies have not described the effect of the downregulation of FT1, which is important to determine if this gene is essential for flowering in the temperate cereals. In Brachypodium FT1 RNAi transgenic lines, the drastic downregulation of FT1 was associated with a non-flowering phenotype, even after the plants were vernalized for two weeks. This result indicates that FT1 is essential for flowering in this Brachypodium line under the conditions tested in this study.
Rice FT1 homologs also seem to be essential for flowering [42]. In this species, there are two duplicated FT1 orthologs, Hd3a and the Rice Flowering Locus T1 (RFT1) that share 90% identity at the protein level, suggestive of a relatively recent duplication in the rice lineage [42]. The simultaneous silencing of Hd3a and RFT1 by RNAi results in non-flowering rice plants, even when grown in an inductive SD photoperiod [42]. In contrast, non-functional mutations in all six FT-like genes in Arabidopsis delay, but do not abolish flowering [43]. These studies in rice and Arabidopsis indicate that the importance of FT-like genes in flowering regulation carries across species. Our Brachypodium and wheat results support this conclusion. While FT1 seems to be essential for flowering in Brachypodium, in wheat the downregulation by RNAi or disruptive mutation in the coding regions result only in delayed flowering.
In wheat, most FT1 RNAi transgenic plants (10 out of 13 T0 lines) flowered only two to four weeks later than the wild type Bobwhite plants (Table 2) and were able to set seeds. Among the three extremely late or non-flowering wheat FT1 RNAi transgenic lines only one set seed (line ‘1965’). A progeny test of this line showed that the derived T1 progeny flowered 10 to 20 days later than the wild type, similar to the other transgenic lines (Table 2). This result suggests that other factors (e.g. tissue culture) may have contributed to the later flowering time of the ‘1965’ T0 plant relative to its T1 progeny. The other two late flowering wheat plants (lines ‘2547’ and ‘2548’) failed to produce seeds and, therefore, it was not possible to verify whether their unusually late flowering phenotype was linked to the FT1 RNAi transgene.
The stronger effect of the FT1 RNAi transgene on flowering time in Brachypodium relative to wheat was correlated with a stronger downregulation of FT1 in the Brachypodium transgenic lines (<1% of wild type) than in wheat (∼26% of wild type). We currently do not know if this is caused by differences in the efficiency of the RNAi constructs, or by the different genetic backgrounds. The Bd21-3 line selected for Brachypodium transformation has a facultative winter growth habit [40] whereas the Bobwhite genotype used in wheat transformation has a spring growth habit. The Vrn-A1, Vrn-B1 and Vrn-D1 alleles present in Bobwhite are dominant for early flowering and epistatic to the winter alleles of other vernalization genes. These VRN1 alleles may contribute, directly or indirectly, to the relatively higher FT1 transcript levels observed in the wheat RNAi lines than in the Brachypodium RNAi lines.
The tetraploid line Kronos used to generate the ft1 TILLING mutants also carries a Vrn-A1 allele that is dominant for spring growth habit and epistatic to other vernalization genes [44]. This may contribute to the limited effect of the ft-A1/ft-B1 263 double mutant, which showed a flowering delay (25 days) similar to that observed in the FT1 RNAi transgenic hexaploid wheat plants. Unfortunately, we were unable to find a truncation mutation for the FT-B1 homoeolog and therefore, we cannot rule out the possibility that the selected ft-B1 263 mutant has some residual ability to induce flowering.
Triple ppd1-null mutants in the photoperiod sensitive hexaploid spring variety ‘Paragon’, which have almost undetectable FT1 transcript levels, flower approximately 30 days later than their wild type control [45]. This delay in flowering is similar to that observed in the ft-A1/ft-B1 263 double mutant and in the Bobwhite FT1 RNAi transgenic lines. Taken together, these results support the hypothesis that FT1 is not essential for flowering in spring wheat. Even if other genes can induce flowering in wheat, our results demonstrate that FT1 still plays a critical role in the timely induction of reproductive development in both Brachypodium and wheat.
Changes Induced by FT1 are Associated with Changes in Other FT-like Genes
FT-like genes FT2, FT3, FT4, FT5 and FT6 are the closest paralogs of FT1 in the temperate grasses [12]. Among these, FT2 is the most similar to FT1 (>75% protein identity), and is part of a duplication that occurred before the divergence of the rice and wheat lineages (Figure 5). The FT3 and FT5 clusters (80 to 84% protein identity) appear to have arisen from a more recent duplication. Wheat and barley FT3 and FT5 proteins form well supported clusters, but their relationship with the Brachypodium homologs is unclear. This suggests that this duplication occurred prior to the divergence of wheat and barley, but close to the time of divergence between the Brachypodium and the Triticeae lineages. A separate cluster includes two groups of wheat, Brachypodium and barley paralogs; the FT4 group that is related to rice OsFTL12 and the FT6 group that is related to rice OsFTL13 (Figure 5). The FT4 and FT6 paralogs have a similar level of divergence (71 to 75% protein identity) to that between the FT1 and FT2 paralogs.
Downregulation of FT1 in the FT1 RNAi transgenic plants in Brachypodium and wheat results in a significant downregulation of FT2, while upregulation of FT1 in FT1 HOPE wheat plants results in a significant upregulation of FT2. These results suggest that, directly or indirectly, FT2 transcription is regulated by FT1. Overexpression of FT2 homologs in Brachypodium [25] and rice (OsFTL1) [46] results in extremely early flowering similar to the results observed for FT1. These results suggest that FT1 and FT2 paralogs have both retained the ability to induce flowering. Therefore, the changes in FT2 transcript levels observed in the FT1 RNAi and FT1 HOPE transgenic plants may also contribute to the observed differences in flowering time.
Significant changes in the expression of other FT-like genes were detected in the FT1 RNAi and FT1 HOPE transgenic plants, but these effects were less consistent across genotypes and environmental conditions. The association between reduced FT1 transcript levels in the FT1 RNAi lines and the downregulation of FT4 was significant only in Brachypodium, whereas the association with the downregulation of FT5 was significant only in wheat. While the effect of FT1 on FT3 expression was significant in both LD and SD grown FT1 HOPE transgenic plants, the transcript levels of wheat FT3 were 25-fold higher under SD than under LD (Figure 8). This has also been reported previously in barley, where FT3 is a candidate gene for a major QTL affecting barley flowering under SD (Ppd-H2) [12]. Overexpression of barley FT3 in rice also accelerates flowering [47]. However, this effect is significantly smaller than the acceleration of flowering time in rice transgenic lines transformed with the barley FT1 and FT2 genes and is modulated by photoperiod [47]. These results suggest that FT3 may play a role distinct from that of FT1 and FT2 in the regulation of flowering time.
No significant differences in FT4, FT5 or FT6 transcript levels were detected between Jagger and the Jagger FT1 HOPE transgenic line, but the transcript levels of these three genes were higher under LD than under SD in the wild type Jagger plants suggesting some role in the photoperiodic response (Figure 8). The roles of FT4, FT5 and FT6 and their rice homologs remain poorly understood.
The observed reduction in the expression of FT-like genes in the FT1 RNAi transgenic plants is unlikely to be the result of a direct targeting of the RNAi construct, because the region of FT1 selected as target for RNAi did not share any stretches of more than 18 identical nucleotides with other FT-like genes. A continuous stretch of at least 21 identical nucleotides between the trigger and a target gene is required to produce efficient silencing [48]. Therefore, the transcriptional changes observed in other FT-like genes in the leaves of the transgenic plants are likely a result of the changes in FT1 or of the developmental changes induced by FT1. Additional studies will be required to validate the effect of FT1 on the expression of other FT-like genes.
In rice, co-silencing of the FT1 homologs Hd3a and RFT1 also resulted in the downregulation of the FT2 homolog OsFTL1, but showed no effect on the expression of the rice homologs of FT3 and FT5 (OsFTL10) or FT4 (OsFTL12) [42]. Therefore, the observed changes in FT4 and FT5 transcription could be species specific, or be affected by different developmental stages of the plants in the different experiments.
The fact that FT1 interacts with the bZIP transcription factors FDL2 and FDL6 (which can bind the VRN1 promoter), while FT2 interacts with FDL13 (which cannot bind the VRN1 promoter) suggests that FT1 and FT2 may have different gene targets [21]. This observation is consistent with the functional divergence observed for the FT1 and FT2 orthologs in rice (Hd3a and OsFTL1). Whereas overexpression of Hd3a in rice results in an early flowering phenotype [49], overexpression of OsFTL1 produces a more complex phenotype including elongation of internodes, loss of apical dominance, and formation of a terminal tissue at the apical meristem [46].
In rice, Hd3a forms a hexameric protein complex comprised of two 14-3-3 proteins, two FD-like proteins and two Hd3a proteins, designated as the florigen activation complex (FAC) [50]. This complex has been shown to bind the OsMADS15 promoter (a homologue of VRN1) and induce flowering. Different combinations of FT-like, FD-like and 14-3-3 proteins can theoretically form a large number of possible different FACs, each varying in their constituent components. This system may provide flexibility to translate signals carried by FT-like mobile protein into tissue and developmental stage specific responses by modulating the expression of FD-like or 14-3-3 proteins interacting proteins in those tissues or developmental stages [51]. Our understanding of these complex interactions is in its infancy, but the results presented here suggest that changes in FT1 are associated with changes in the expression of other FT-like genes, which may help to coordinate the complex responses required during the transition from the vegetative to the reproductive phase. It would be interesting to test if the alteration of the expression of other FT-like genes affects the expression of FT1, or if this is a unidirectional hierarchical regulatory system in which FT1 initiates a cascade of changes that affects the expression of the other members of the FT-like family.
TILLING Mutants of FT-A1 and FT-B1 Homoeologs Provide New Variability to Engineer Wheat Flowering Time
The ft-A1 mutant has a premature stop codon that eliminates more than half of the encoded protein whereas the ft-B1 263 has an amino acid substitution within the conserved PEBP domain. Therefore, we initially expected the ft-A1 mutant to exhibit a larger effect on flowering time than the ft-B1 263 mutant. However, we observed the opposite; a significantly larger delay in flowering was associated with the ft-B1 263 mutants than with the ft-A1 mutant. This result suggests that the FT-B1 homoeolog has a more important role in the regulation of flowering in tetraploid wheat than the FT-A1 homoeolog.
The FT-A1 and FT-B1 proteins are identical, so differences in their transport efficiency or in their ability to induce flowering are unlikely to contribute to the observed differences in heading time. By contrast, the higher transcript levels of FT-B1 relative to FT-A1, correlates well with the larger effect of FT-B1 relative to FT-A1 on wheat heading time, providing a simple explanation for the observed phenotypic effects. Similar differences have been described for hexaploid wheat, where FT-B1 exhibits much higher expression levels than either the A or D homoeologs [15]. Although the ft-A1 mutation showed no effect on flowering time, it was associated with a delay in heading time of 6.7 days in plants homozygous for the ft-B1 263 mutation. This suggests that FT-A1 is a hypomorphic allele, but that it still retains some ability to induce flowering under LD.
The slight differences in heading time between the two different ft-B1mutants and among the different combination of ft-A1 and ft-B1 alleles suggest that different combinations of FT1 homoeologs (both mutant and natural alleles) can be used to precisely regulate heading time in wheat. This will become more important as wheat breeders try to adjust heading time of current wheat varieties in environments affected by global climate change.
Funding Statement
This research was supported by the National Natural Science Foundation of China (31110103917, 30871323) and the Natural Science Foundation of Shandong Province (JQ201107). Dr. Dubcovsky and Rebecca Nitcher acknowledge support from the National Research Initiative (grants 2011-67013-30077 and 2011-68002-30029) from the USDA National Institute of Food and Agriculture, and by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation grant GBMF3031. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Brambilla V, Fornara F (2013) Molecular control of flowering in response to day length in rice. J Integr Plant Biol 55: 410–418. [DOI] [PubMed] [Google Scholar]
- 2. Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639. [DOI] [PubMed] [Google Scholar]
- 3. Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12: 178–184. [DOI] [PubMed] [Google Scholar]
- 4. Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, et al. (2003) Positional cloning of the wheat vernalization gene VRN1 . Proc Natl Acad Sci USA 100: 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100: 13099–13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, et al. (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yan L, Fu D, Li C, Blechl A, Tranquilli G, et al. (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT . Proc Natl Acad Sci USA 103: 19581–19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoshida T, Nishida H, Zhu J, Nitcher R, Distelfeld A, et al. (2010) Vrn-D4 is a vernalization gene located on the centromeric region of chromosome 5D in hexaploid wheat. Theor Appl Genet 120: 543–552. [DOI] [PubMed] [Google Scholar]
- 9. Kippes N, Zhu J, Chen A, Vanzetti L, Lukaszewski A, et al. (2014) Fine mapping and epistatic interactions of the vernalization gene VRN-D4 in hexaploid wheat. Mol Genet Genomics 289: 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, et al. (2006) Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2 . Plant Mol Biol 60: 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trevaskis B, Hemming MN, Peacock WJ, Dennis ES (2006) HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol 140: 1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Faure S, Higgins J, Turner A, Laurie DA (2007) The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen A, Dubcovsky J (2012) Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet 8: e1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034. [DOI] [PubMed] [Google Scholar]
- 15. Shaw LM, Turner AS, Laurie DA (2012) The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum). Plant J 71: 71–84. [DOI] [PubMed] [Google Scholar]
- 16. Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115: 721–733. [DOI] [PubMed] [Google Scholar]
- 17. Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis . Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- 18. Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594. [DOI] [PubMed] [Google Scholar]
- 19. Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036. [DOI] [PubMed] [Google Scholar]
- 20. Lin M-K, Belanger H, Lee Y-J, Varkonyi-Gasic E, Taoka K-I, et al. (2007) FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19: 1488–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li C, Dubcovsky J (2008) Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J 55: 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pearce S, Vanzetti LS, Dubcovsky J (2013) Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1 . Plant Physiol 163: 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwartz C, Doyle M, Manzaneda A, Rey P, Mitchell-Olds T, et al. (2010) Natural variation of flowering time and vernalization responsiveness in Brachypodium distachyon . BioEnergy Res 3: 38–46. [Google Scholar]
- 24. Shimada S, Ogawa T, Kitagawa S, Suzuki T, Ikari C, et al. (2009) A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. Plant J. 58: 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu L, Liu D, Wu J, Zhang R, Qin Z, et al. (2013) Regulation of FLOWERING LOCUS T by a MicroRNA in Brachypodium distachyon . Plant Cell 25: 4363–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ream T, Woods D, Schwartz C, Sanabria C, Mahoy J, et al. (2013) Interaction of photoperiod and vernalization determine flowering time of Brachypodium distachyon. Plant Physiol. doi:10.1104/pp.113.232678. [DOI] [PMC free article] [PubMed]
- 27. Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, et al. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41. [DOI] [PubMed] [Google Scholar]
- 28. Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495. [DOI] [PubMed] [Google Scholar]
- 29. Bragg JN, Wu J, Gordon SP, Guttman ME, Thilmony R, et al. (2012) Generation and characterization of the Western Regional Research Center Brachypodium T-DNA insertional mutant collection. PLoS ONE 7: e41916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weeks JT, Anderson OD, Blechl AE (1993) Rapid production of multiple independent lines of fertile transgenic wheat (Triticum aestivum). Plant Physiol 102: 1077–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, et al. (2009) A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol 9: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falconer DS (1964) Introduction to quantitative genetics. Edinburgh: Oliver & Boyd.
- 33. Fu D, Dunbar M, Dubcovsky J (2007) Wheat VIN3-like PHD finger genes are up-regulated by vernalization. Mol Genet Genomics 277: 301–313. [DOI] [PubMed] [Google Scholar]
- 34. Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314: 1298–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peterson R, Slovin JP, Chen C (2010) A simplified method for differential staining of aborted and non-aborted pollen grains. Intl J Plant Biology 1: e13. [Google Scholar]
- 36. Henikoff S, Henikoff JG (1992) Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 89: 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sawa M, Kay SA (2011) GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana . Proc Natl Acad Sci USA 108: 11698–11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J (2005) Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol 138: 2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oliver SN, Finnegan EJ, Dennis ES, Peacock WJ, Trevaskis B (2009) Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc Natl Acad Sci USA 106: 8386–8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ream TS, Woods DP, Amasino RM (2012) The molecular basis of vernalization in different plant groups. Cold Spring Harb Sym 77: 105–115. [DOI] [PubMed] [Google Scholar]
- 41. Nitcher R, Distelfeld A, Tan C, Yan L, Dubcovsky J (2013) Increased copy number at the HvFT1 locus is associated with accelerated flowering time in barley. Mol Genet Genomics 288: 261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135: 767–774. [DOI] [PubMed] [Google Scholar]
- 43. Kim W, Park TI, Yoo SJ, Jun AR, Ahn JH (2013) Generation and analysis of a complete mutant set for the Arabidopsis FT/TFL1 family shows specific effects on thermo-sensitive flowering regulation. J Exp Bot 64: 1715–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fu D, Szücs P, Yan L, Helguera M, Skinner JS, et al. (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics 273: 54–65. [DOI] [PubMed] [Google Scholar]
- 45. Shaw LM, Turner AS, Herry L, Griffiths S, Laurie DA (2013) Mutant alleles of Photoperiod-1 in wheat (Triticum aestivum L.) that confer a late flowering phenotype in long days. PLoS ONE 8: e79459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, et al. (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Gene Dev 16: 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kikuchi R, Kawahigashi H, Ando T, Tonooka T, Handa H (2009) Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol 149: 1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fu D, Uauy C, Blechl A, Dubcovsky J (2007) RNA interference for wheat functional gene analysis. Transgenic Res 16: 689–701. [DOI] [PubMed] [Google Scholar]
- 49. Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, et al. (2002) Hd3a, a rice ortholog of the Arabidopsis FT Gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105. [DOI] [PubMed] [Google Scholar]
- 50. Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, et al. (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335. [DOI] [PubMed] [Google Scholar]
- 51. Taoka K, Ohki I, Tsuji H, Kojima C, Shimamoto K (2013) Structure and function of florigen and the receptor complex. Trends Plant Sci 18: 287–294. [DOI] [PubMed] [Google Scholar]
- 52. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W (2011) Cloning and functional analysis of VRT2-like Genes in Brachypodium distachyon L. Tai’an, China: Shandong Agricultural University. 76 p.
- 54. Krasileva KV, Buffalo V, Bailey P, Pearce S, Ayling S, et al. (2013) Separating homeologs by phasing in the tetraploid wheat transcriptome. Genome Biol 14: R66. [DOI] [PMC free article] [PubMed] [Google Scholar]