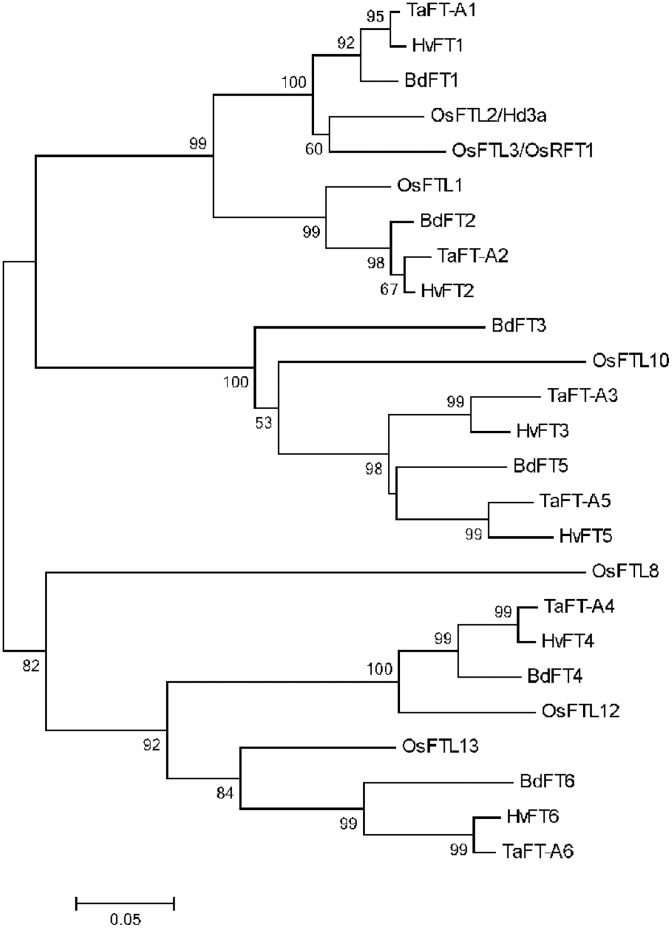
Figure 5. Phylogenetic analysis of FT-like genes in temperate grasses.
Phylogenetic analysis was performed using the full-length proteins. A neighbor-joining tree was constructed using pairwise deletions and 1,000 bootstrap iterations with the program MEGA 5.0 [52]. The scale bar 0.05 represents 5% base substitution. Bootstrap numbers larger than 50 are shown in the respective nodes. To simplify the tree, only the wheat A-genome homoeologs of wheat were included. Accession information: BdFT1 (Bradi1g48830), BdFT2 (Bradi2g07070), BdFT3 (Bradi2g49795), BdFT4 (Bradi1g38150), BdFT5 (Bradi2g19670), BdFT6 (Bradi3g08890), HvFT1 (DQ100327), HvFT2 (DQ297407), HvFT3 (DQ411319), HvFT4 (DQ411320), HvFT5 (EF012202), HvFT6 (morex_contig_54196), OsFTL1 (Os01g11940), OsFTL2/Hd3a (Os06g06320), OsFTL3/RFT1 (Os06g06300), OsFTL8 (Os01g10590), OsFTL10 (Os05g44180), OsFTL12 (Os06g35940), OsFTL13 (Os02g13830), TaFT-A1 (CD881060), TaFT-A2 (BT009051), TaFT-A3 (IWGSC_1AL_913428), TaFT-A4 (IWGSC_2AS_5252557), TaFT-A5 (IWGSC_5AL_2803506), TaFT-A6 (IWGSC_6AS_4388307). HvFT6 sequence is from the International Barley Sequencing Consortium (IBSC, http://webblast.ipk-gatersleben.de/barley/).