Abstract
Microsomal cytochrome b5 plays a key role in the oxidation of a variety of exogenous and endogenous compounds, including drugs, fatty acids, cholesterol and steroid hormones. To better understand its functional properties in a membrane mimic environment, we carried out high-resolution solution NMR studies. Here we report resonance assignments for full-length rabbit cytochrome b5 embedded in DPC (dodecylphosphocholine) micelles.
Keywords: cytochrome b5, membrane protein, Heteronuclear NMR
Biological context
Cytochromes b5 (cytsb5) are ubiquitous electron transport proteins found in plants, animals, fungi and prokaryotic organisms. In eukaryotes, cytsb5 exist as membrane-anchored proteins found either in the endoplasmic reticulum (ER) or in the outer mitochondrial membrane (Schenkman et al 2003). In animal erythrocytes and prokaryotes, cytsb5 are mostly found in a water-soluble form that lacks the C-terminal transmembrane domain (Dürr et al 2007; Vergeres et al, 1995). The isoform of the full-length cytb5, which resides on the cytoplasmic side of the ER membrane (referred to as microsomal cytb5), is a ~16-kDa (134 amino acids), predominantly an acidic membrane protein consisting of two separate domains: a large, N-terminal, cytosolic heme-containing soluble domain (~94 amino acids) that includes the binding site for its redox partners and a C-terminal hydrophobic transmembrane domain (~23 amino acids). These two domains are connected by a proline containing hinge region of ~14 residues referred to as the linker (Clarke et al. 2004). Cytb5 contains a type B heme, which is located in the hydrophobic core of the soluble cytosolic domain, with the highly conserved His68 and His44 coordinating the heme iron as the 5th and 6th ligands (Lederer 1994). Microsomal cytsb5 participate in a number of key reactions, including fatty acid desaturation (Dürr et al. 2007), the biosynthesis of cholesterol (Schenkman et al. 2003) and sex hormones (Kominami et al. 1992), and the hydroxylation of N-acetyl-neuraminic acid (Takematsu et al. 1994).
High-resolution structures have been determined for the cytosolic, heme-binding domain of truncated, microsomal cytb5 from solution NMR and X-ray crystallography (Banci et al, 2000; Nunez et al. 2010; Durley et al. 1996). Solution NMR studies have reported the dynamics of the truncated cytb5 in solution (Arnesano et al, 2000; Banci et al, 2001). Solid-state NMR studies on magnetically-aligned bicelles containing the full-length rabbit cytb5 reported the fast dynamics (microsecond dynamics) of the soluble domain, slow mobility (millisecond time scale) of the transmembrane domain, and the topology of the transmembrane domain (Dürr et al. 2007b; Xu et al. 2008; Soong et al. 2010; Xu et al. 2010). The tilt angle of the transmembrane helix of cytb5 and its membrane insertion process has been investigated using solid-state NMR and sum frequency generation experiments in phospholipid bilayers (Dürr et al. 2007b; Nguyen et al. 2010). However, the lack of any high-resolution structural data for the full-length cytb5 makes it particularly difficult to establish the molecular mechanism of electron transfer upon its interaction with the monooxygenase, cytochrome P450 (cytP450) (Schenkman et al. 2003; Dürr et al. 2007).
Thus, to elucidate the structure of full-length microsomal cytb5 in a membrane mimetic two- and three-dimensional heteronuclear (13C, 15N) NMR spectroscopy were performed (Ahuja et al. 2013). Here we present the assignment of 1H, 13C, and 15N resonances for cytb5 protein embedded in DPC micelles.
Methods and experiments
C41 cells were purchased from Lucigen (Middleton, MI). U-13C, 15N and 2H CELTONE rich medium, 15N-CELTONE rich media, 13C, 15N-CELTONE rich media, 2H-dodecylphosphocholine (DPC-D38), 13C-glucose, 15N-ammonium sulfate and D2O were purchased from Cambridge Isotope Laboratories (Andover, MA). Resins and buffer components were purchased from Sigma-Aldrich. Glycerol for NMR experiments was purchased from Sigma-Aldrich and Roche Applied Science. The NMR samples were placed into 5 mm symmetrical D2O-matched Shigemi NMR microtubes (Shigemi, Inc, Alison Park, PA).
Wild-type rabbit, full-length cytochrome b5 was overexpressed and purified using the protocols described previously (Xu et al. 2008). U-15N cytb5, U-15N, 13C cytb5 and U-15N, 13C, 2H cytb5 were expressed using Celtone-N, Celtone-CN and Celtone-DCN complete media, respectively, with additional supplements as described in reference (Nunez et al. 2010 and Ahuja et al. 2013). Purification of cytb5 was performed as described elsewhere (Mulrooney et al. 2000). Each purified protein exhibited a single band on an SDS PAGE gel.
NMR Spectroscopy
All NMR experiments were performed on a Bruker Avance 900 MHz four-channel NMR spectrometer equipped with an x,y,z axis PFG 5 mm TCI cryoprobe. NMR sample was prepared by reconstituting 0.1 – 0.5 mM cytb5 in 100 mM potassium phosphate buffer containing 5 % deuterated glycerol in the presence of 45 mM perdeuterated DPC (DPC-D38) at pH 7.4.
Two-dimensional TROSY-based 1H-15N (Pervushin et al. 1997) and 1H-13C heteronuclear single-quantum coherence (HSQC) spectra and three-dimensional TROSY based (3D) HNCA, HNCO, HNCACB, HN(CA)CO, HN(CO)CA, 15N- edited TOCSY-HSQC (Sattler et al. 1999) were collected for the backbone chemical shifts assignments. 15N, 13C and 2H labeled protein was used for all the triple resonance backbone NMR experiments. For 15N-HSQC-NOESY and 13C-HSQC-NOESY experiments, uniformly 15N and 13C labeled cytb5 embedded in DPC-D38 was used. The 3D-NOESY (with mixing times 80 and 100 ms) experiments were used to confirm the chemical shift assignment in addition to obtaining intra and inter-residue NOEs. All aromatic side chain protons and carbon atoms were assigned using 2D-NOESY and 3D-NOESY experiments. Time to time, several 2D TROSY 1H-15N HSQC spectra were recorded to monitor sample stability. The proton chemical shifts were referenced to the methyl signal of 2,2-dimethyl-2-silapentane-sulfonic acid (DSS, Cambridge Isotope Laboratories) as an internal chemical shift reference at 0.0 ppm. The 13C and 15N chemical shifts were referenced indirectly to DSS (Harris et al. 2001). All the above NMR experiments were performed at 25 C.
All NMR spectra were processed by either NMRPipe (Delaglio et al. 1995) or Topspin 2.0 (Bruker) and analyzed using Sparky (Kneller et al. 1993).
Assignment and Data Deposition
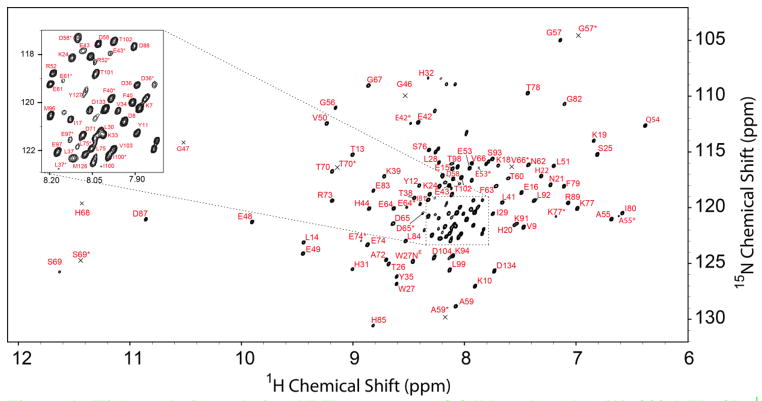
The assigned 1H-15N TROSY-HSQC spectrum for full-length cytb5 is shown in Fig. 1. Using standard three-dimensional solution NMR experiments, NMR resonance assignment was achieved for 88.5% of the backbone and side chain atoms of residues from the soluble heme-binding domain of full-length cytb5 (2D spectral strips illustrating resonance assignments are shown in Figures 2 and 3). Besides three prolines, the unassigned residues in the heme-binding domain of cytb5 include M1-D6, S23, K33, K91, K94, I100, S105, due to their flexibility and rapid solvent exchange with their amide protons. Ambiguous assignments were made for the residues N106, A124, M126, Y127, R128, D133 and D134 due to broad and overlapped peaks in all 3D triple resonance and 15N/13C-edited 3D- HSQC-NOESY spectra. No backbone assignments were made for the transmembrane domain residues S107-V123, L125, L129, Y130, M131 and A132 as no resonance peaks were identified for these residues in the 1H-15N TROSY-HSQC spectrum of cytb5. The restricted slow (millisecond or slower) motion of the transmembrane domain of cytb5 incorporated in DPC micelles causes significant broadening of the transmembrane domain resonances due to fast spin-spin relaxation. As described in our previous work, a 1H-15N-HMQC spectrum recorded under magic angle spinning (2.5 kHz) on a selectively 15N-alanine labeled sample of cytb5 incorporated in DPC micelles displayed broad resonances for the backbone amide-NHs of the four alanines present in the transmembrane domain of cytb5, along with narrow resonances for the alanines in the soluble domain (Dürr et al. 2007b; Ahuja et al. 2013, Fig. 4). Hence, static solid-state NMR experiments were performed on uniformly 15N-labeled full-length cytb5 incorporated in magnetically-aligned bicelles - composed of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) lipids in a 3.5:1 molar ratio - to obtain the structure of the transmembrane domain of cytb5 (Ahuja et al. 2013).
Figure 1. High-resolution solution NMR spectrum of full-length cytb5.
(A) 900 MHz 2D 1H-15N-TROSY-HSQC spectrum of a uniformly 15N, 13C and 2H- labeled full-length mammalian cytb5 in NMR buffer at pH 7.4 and 45 mM DPC micelles. The assignments for resolved backbone residues are labeled with one letter amino acid code and residue number. The peaks marked by an asterisk (*) indicate the amino acid residue assignment from the minor population of cytb5 isomer. An expansion of the crowded region of the 2D 1H-15N-TROSY-HSQC spectrum is inserted in the figure for clarity. The low intensity peaks are marked as ‘x’ with residue assignment.
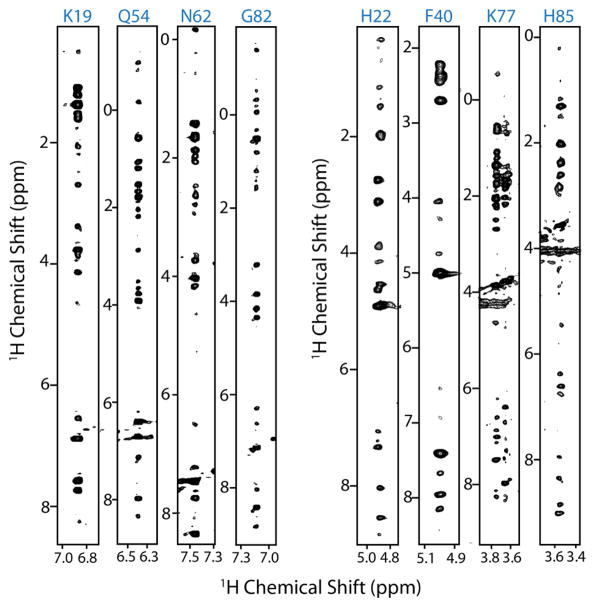
Figure 2.
1H-1H planes of eight different residues extracted from a 3D 15N-edited HSQC-NOESY (mixing time 100 ms)recorded on a fully protonated uniformly 13C and 15N labeled cytb5 in DPC micelles.
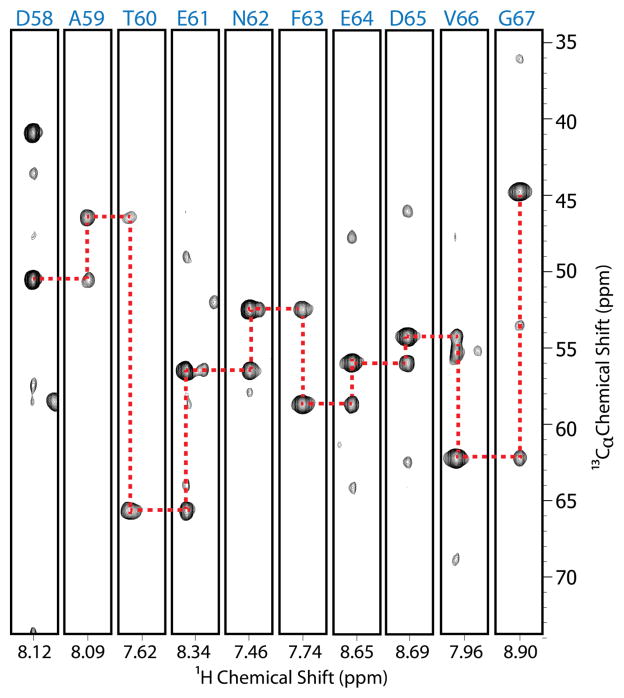
Figure 3.
Strips from HNCA spectrum used to make backbone assignments. Each strip is labeled on top by the amino acid whose NH was detected and at the bottom with the 1HN chemical shift.
An inspection of the 1H-15N-TROSY-HSQC spectrum (Fig 1) of cytb5 revealed two or more NMR resonances (marked as *) for many of the residues. These two sets of NMR resonances originate from the two isomers (major and minor) of cytb5 that differ by a 180° rotation of the heme plane about the axis that cuts through the meso-carbon atoms α and γ (Banci et al. 2000; Zhang et al. 2004). The ratio of the populations of the two isomers can be calculated by determining the peak intensity ratio (here in the 1H-15N-TROSY-HSQC spectrum of cytb5) for identical residues in the two isomeric forms. The major/minor isomer ratio in our study for the full-length rabbit cytb5 was determined to be about 6.6:1 which is similar to the previously obtained 5:1 ratio for truncated rabbit cytb5 (Banci et al. 2000) and nearly identical to the isomer ratio of 6.5:1 for truncated bovine cytb5 (Zhang et al. 2004).
The analysis of 15N-HSQC-NOESY and 13C-HSQC-NOESY reveals that the soluble, heme-binding domain (M1-D89) of cytb5 consists of six α-helices, five β-strands. The linker region (S90-D104) was found to be completely unstructured.
A list of the 1H, 13C and 15N chemical shift values has been deposited into the BioMagResBank (http://www.bmrb.wisc.edu) under accession number BMRB - 18919. Although backbone assignments were done for the resonance peaks of both the major and minor isomers, all the reported assignments in BMRB are only for the major isomer of the ferric full-length microsomal cytb5.
Acknowledgments
This research was supported by funds from NIH (GM084018 and GM095640 to A. R., and GM35533 to L. W.). We would like to thank Dr. Russell Pandian and Rui Huang for help with figures.
References
- Ahuja S, Jahr N, Im SC, Vivekanandan S, Popovych N, Le Clair SV, Huang R, Soon R, Xu J, Yamamoto K, Nanga RP, Bridges A, Waskell L, Ramamoorthy A. A model of the membrane-bound Cytochrome b5-Cytochrome P450 Complex from NMR and Mutagenesis Data. J Biol Chem. 2013;288:22080–22095. doi: 10.1074/jbc.M112.448225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesano F, Banci L, Bertini I, Koulougliotis D, Monti A. Monitoring mobility in the early steps of unfolding: the case of oxidized cytochrome b5 in the presence of 2 M guanidinium chloride. Biochemistry. 2000;39:7117–30. doi: 10.1021/bi992756k. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Rosato A, Scacchieri S. Solution structure of oxidized microsomal rabbit cytochrome b5 - Factors determining the heterogeneous binding of the heme. Eur J Biochem. 2000;267:755–766. doi: 10.1046/j.1432-1327.2000.01054.x. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Branchini BR, Hajieva P, Spyroulias GA, Turano P. Dimethyl propionate ester heme-containing cytochrome b5: structure and stability. J Biol Inorg Chem. 2001;6:490–503. doi: 10.1007/s007750100217. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Felli IC, Hajieva P, Viezzoli MS. Side chain mobility as monitored by CH-CH cross correlation: the example of cytochrome b5. J Biomol NMR. 2001;20:1–10. doi: 10.1023/a:1011245101351. [DOI] [PubMed] [Google Scholar]
- Clarke TA, Im SC, Bidwai A, Waskell L. The role of the length and sequence of the linker domain of cytochrome b5 in stimulating cytochrome P4502B4 catalysis. J Biol Chem. 2004;279:36809–36818. doi: 10.1074/jbc.M406055200. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPIPE - a multidimensional spectral processing system based on unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Durley RCE, Mathews FS. Refinement and structural analysis of bovine cytochrome b5 at 1.5 angstrom resolution. Acta Crystallogr Sect D-Biol Crystallogr. 1996;52:65–76. doi: 10.1107/S0907444995007827. [DOI] [PubMed] [Google Scholar]
- Dürr UH, Waskell L, Ramamoorthy A. The cytochromes P450 and b5 and their reductases-Promising targets for structural studies by advanced solid-state NMR spectroscopy. Biochim Biophys Acta, Biomembr. 2007a;1768:3235–3259. doi: 10.1016/j.bbamem.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Dürr UH, Yamamoto K, Im SC, Waskell L, Ramamoorthy A. Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b5. J Am Chem Soc. 2007b;129:6670–6671. doi: 10.1021/ja069028m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RK, Becker ED, De Menezes SMC, Goodfellow R, Granger P. NMR nomenclature. Nuclear spin properties and conventions for chemical shifts - (IUPAC recommendations. Pure Appl Chem. 2001;73:1795–1818. doi: 10.1006/snmr.2002.0063. [DOI] [PubMed] [Google Scholar]
- Kneller DG, Kuntz ID. UCSF Sparky - an NMR display, annotation and assignment tool. J Cell Biochem. 1993;53:254–254. [Google Scholar]
- Kominami S, Ogawa N, Morimune R, Huang DY, Takemori S. The role of cytochrome b5 in adrenal microsomal steroidogenesis. J Steroid Biochem. 1992;42:57–64. doi: 10.1016/0960-0760(92)90011-7. [DOI] [PubMed] [Google Scholar]
- Lederer F. The cytochrome b5-fold: an adaptable module. Biochimie. 1994;76:674–692. doi: 10.1016/0300-9084(94)90144-9. [DOI] [PubMed] [Google Scholar]
- Mulrooney SB, Waskell L. High-level expression in Escherichia coli and purification of the membrane-bound form of cytochrome b5. Protein Expr Purif. 2000;19:173–178. doi: 10.1006/prep.2000.1228. [DOI] [PubMed] [Google Scholar]
- Nguyen KT, Soong R, Im S-C, Waskell L, Ramamoorthy A, Chen Z. Probing the Spontaneous Membrane Insertion of a Tail-Anchored Membrane Protein by Sum Frequency Generation. J Am Chem Soc. 2010;132:15112–15115. doi: 10.1021/ja106508f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez M, Guittet E, Pompon D, Van Heijenoort C, Truan G. NMR structure note: oxidized microsomal human cytochrome b5. J Biomol NMR. 2010;47:289–295. doi: 10.1007/s10858-010-9428-6. [DOI] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T-2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc. 1999;34:93–158. [Google Scholar]
- Schenkman JB, Jansson I. The many roles of cytochrome b5. Pharmacol Ther. 2003;97:139–152. doi: 10.1016/s0163-7258(02)00327-3. [DOI] [PubMed] [Google Scholar]
- Soong R, Smith PE, Xu J, Yamamoto K, Im SC, Waskell L, Ramamoorthy A. Proton-evolved local-field solid-state NMR studies of cytochrome b5 embedded in bicelles, revealing both structural and dynamical information. J Am Chem Soc. 2010;132:5779–5788. doi: 10.1021/ja910807e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takematsu H, Kawano T, Koyama S, Kozutsumi Y, Suzuki A, Kawasaki T. Reaction mechanism underlying CMP-N-acetylneuraminic acid hydroxylation in mouse-liver - Formation of a ternary complex of cytochrome b5, CMP-N-acetylneuraminic acid, and a hydroxylation enzyme. J Biochem. 1994;115:381–386. doi: 10.1093/oxfordjournals.jbchem.a124347. [DOI] [PubMed] [Google Scholar]
- Vergeres G, Waskell L. Cytochrome b5, its functions, structure and membrane topology. Biochimie. 1995;77:604–620. doi: 10.1016/0300-9084(96)88176-4. [DOI] [PubMed] [Google Scholar]
- Xu J, Dürr UHN, Im SC, Gan Z, Waskell L, Ramamoorthy A. Bicelle-Enabled Structural Studies on Membrane Protein Cytochrome b5 by Solid-State MAS NMR Spectroscopy. Angew Chem Int Ed. 2008;47:7864–7867. doi: 10.1002/anie.200801338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Soong R, Im SC, Waskell L, Ramamoorthy A. INEPT-Based Separated-Local-Field NMR Spectroscopy: A Unique Approach to Elucidate Side-Chain Dynamics of Membrane-Associated Proteins. J Am Chem Soc. 2010;132:9944–9947. doi: 10.1021/ja103983f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Cao CY, Wang ZQ, Wang YH, Wu HM, Huang ZX. The comparative study on the solution structures of the oxidized bovine microsomal cytochrome b5 and mutant V45H. Prot Sci. 2004;13:2161–2169. doi: 10.1110/ps.04721104. [DOI] [PMC free article] [PubMed] [Google Scholar]