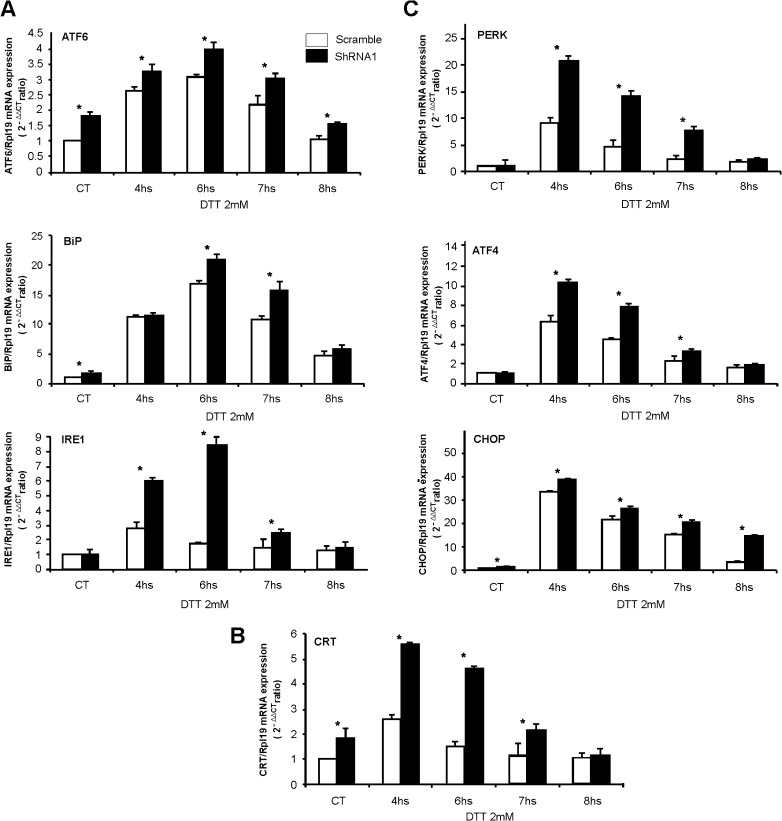
Fig. 4.
Transcriptional effect of RHBDD2 stable silencing on UPR genes under ER stress activation with DTT. Bar charts indicate the level of mRNA expression of each gene under time-dependent ER stress conditions in both scramble and RHBDD2 silenced cells (shRNA1) compared to their respective controls. A significant greater increase of the molecular sensors ATF6 and IRE1 and the chaperone BiP in response to DTT is observed in RHBDD2 silenced cells compared with scramble cells, with the peak of response at 6 h of treatment; additionally, these genes showed high levels of their RNAs in the untreated silenced cells compared with the controls (a). This situation was also evident for the chaperone CRT, but in this case, the peak of response is observed at 4 h (b), as well as observed for PERK, ATF4, and CHOP, which also showed a greater response in silenced cells compared with the scramble (c). Asterisk: Statistically significant differences (p < 0.01)