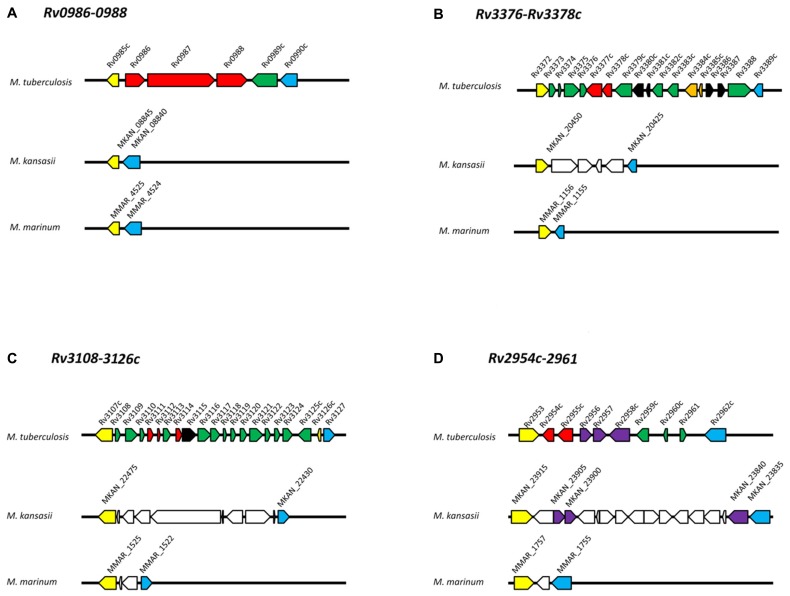
FIGURE 2.
Genomic organization of operons in M. tuberculosis, M. kansasii, and M. marinum. (A) Rv0986-0988; (B) Rv3376-3378c; (C) Rv3108-3126c; (D) Rv2954c-2961. Protein-coding genes are represented by arrows and orthologous genes are indicated by arrows of the same color. Yellow and blue arrows mark the “boundary” of each M. tuberculosis-specific locus. Red arrows indicate genes discussed in this review. Dark green arrows indicate M. tuberculosis genes with no orthologs within the corresponding M. kansasii and M. marinum genomic regions. White arrows in M. kansasii and M. marinum genomes indicate genes present but not orthologs to M. tuberculosis genes. Black arrows indicate transposases; orange arrows indicate toxin-antitoxin genes. Genome organizations for M. tuberculosis, M. kansasii, M. marinum and gene clusters were obtained from the Kyoto Encyclopedia of Genes and Genomes (http://www.kegg.jp/) based on databases available at the Sanger Institute, Tuberculist, and McGill University, respectively. Orthologs were verified by comparing each predicted protein against the H37Rv genome using the BLAST program. Only proteins with >50% coverage, 60% identity and E-value <e-20 were used.