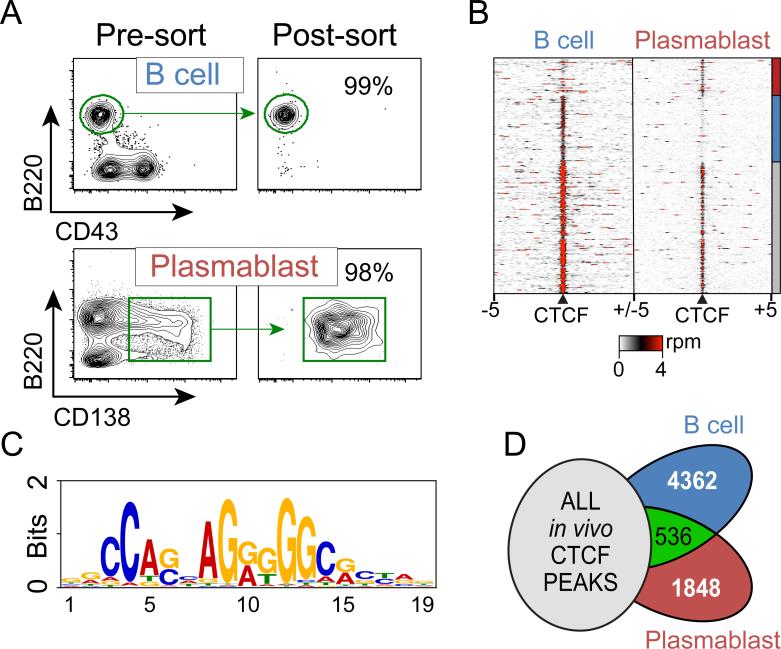
Figure 1. Global CTCF binding profiles in B cells and plasmablasts.
A) A CD43− B cell population was isolated using a magnetic separation procedure and stained with anti-B220-APC and anti-CD43-FITC. 99% of the cells stained positive for the B cell marker B220. Mouse plasmablasts were induced following injection of 50 μg LPS retro-orbitally into 6-week-old C57Bl/6 mice. Splenocytes were isolated from the LPS-induced mice and stained with anit-CD138-PE and anti-B220-APC. CD138+B220int plasmablasts were isolated to high purity by FACS. B) Heat map representing CTCF ChIP-seq read density at all CTCF sites identified in B cells and plasmablasts. Each row represents 5 kb surrounding a CTCF site. Rows are clustered and annotated with a bar on the right to depict shared and cell-type specific CTCF sites. rpm – reads per million. C) Consensus sequence of core homology elements derived from the sequences of CTCF binding sites from B cells and plasmablasts as identified by the MEME-ChIP software (51). D) Venn diagram showing the overlap of in vivo derived CTCF sites identified by the ENCODE Consortium (Supplemental Table 1)(41) and B cells and plasmablasts. 4,362 and 1,848 CTCF sites were specific to B cells and plasmablasts respectively.