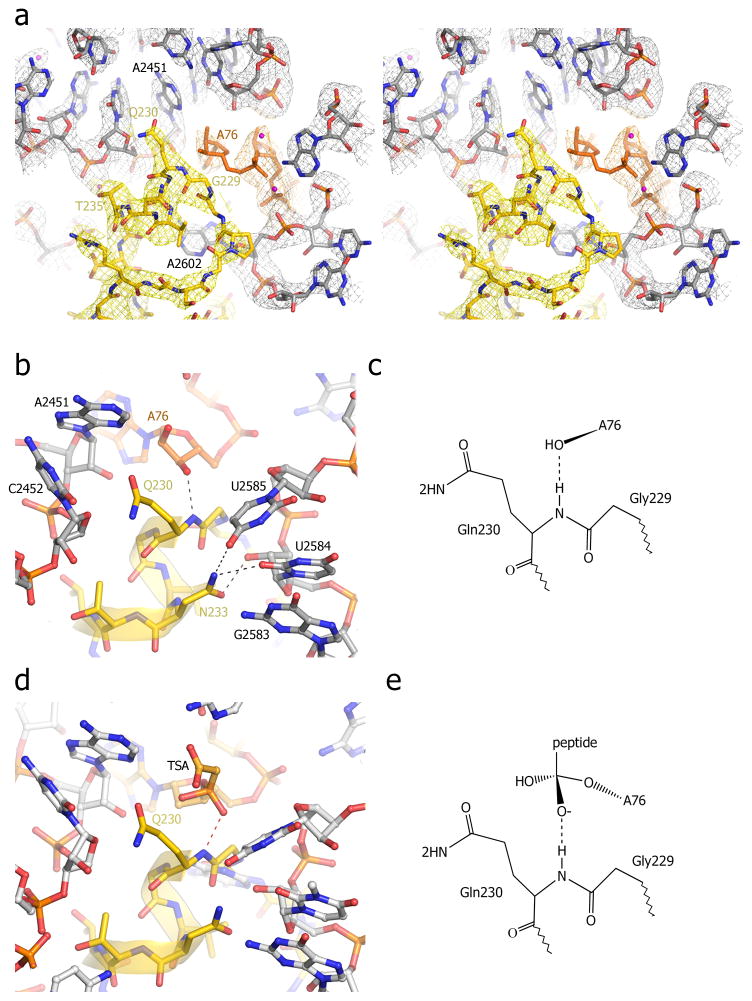
Fig. 3. Interactions of the universally conserved GGQ motif in the peptidyl transferase center (PTC) of the termination complex.
(a) Stereo view of σA-weighted 3Fo-2Fc electron density map for the PTC. Density for RF1 (yellow), P-tRNA (orange) and 23S rRNA (grey) was contoured at 1.7 σ. (b) Interaction of the backbone amide nitrogen of the universally conserved Gln230 with the 3′-OH of A76 of P-site tRNA. (c) Model for proposed product stabilization of the peptide release reaction by H-bonding between the main-chain amide nitrogen of Gln230 and the 3′-OH of A76. (d) Superposition of the structure of the peptidyl-transferase transition-state analog (TSA, orange) complexed with the 50S subunit (grey)[55] on the structure of the termination complex (this work). The main-chain amide nitrogen of Gln230 is positioned to form a hydrogen bond with the oxyanion of the TSA. (e) Model for potential transition-state stabilization of the peptide release reaction by H-bonding between RF1 and the TSA. From ref. [16].