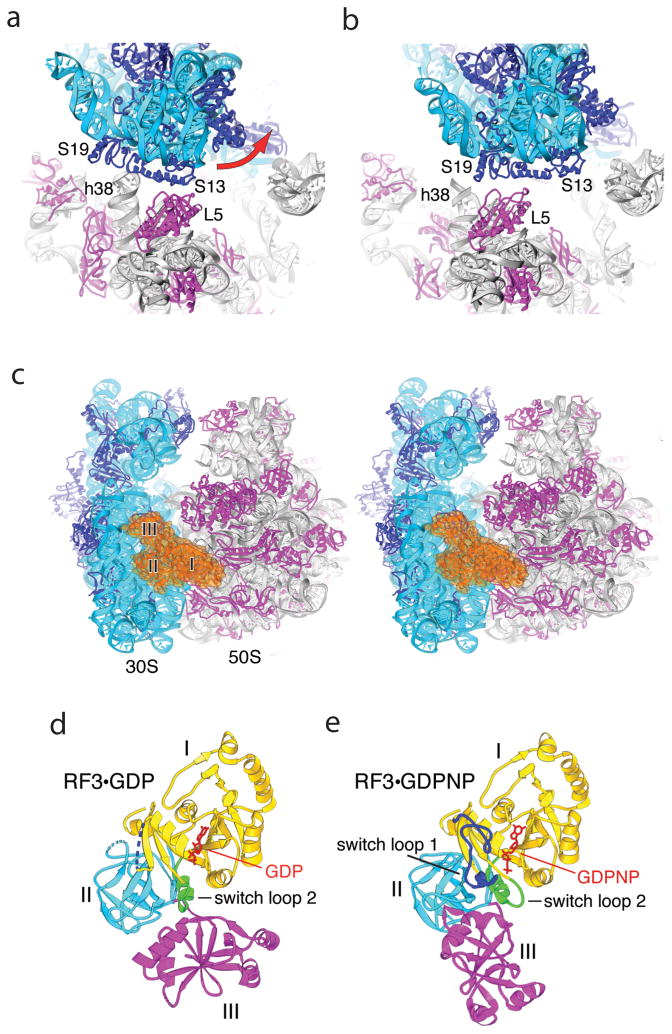
Fig. 4. Effects of binding RF3 to the 70S ribosome.
(a,b) Large-scale rotation of the 30S subunit head induced by RF3 in the E. coli complex [43]. (a) In the classical-state ribosome [17], bridge B1a is formed between protein S13 and helix 38 of 23S rRNA, and B1b between S13 and L5. (b) In the RF3 complex, a large-scale (14°) counter-clockwise rotation of the 30S head results in disruption of bridges B1a and B1b; a new bridge R1b is formed between proteins S13 and S19 from the 30S subunit and L5 from the 50S subunit, involving a 34 Å displacement of the intersubunit contacts. (c) Stereo view of RF3 (orange) bound at the entrance to the interface cavity of the 70S ribosome. (d,e) Comparison of crystal structures of (d) free RF3·GDP [42] and (e) ribosome-bound RF3·GDPNP. A large-scale rotational movement of domain III (magenta) and smaller movements in domains I (yellow) and II (cyan) can be seen. Disordered segments in RF3·GDP are shown as dotted lines. Rearranged segments in RF3·GDPNP containing switch loops I and II are shown in blue and green, respectively. GDP and GDPNP are shown in red. From ref. [43].