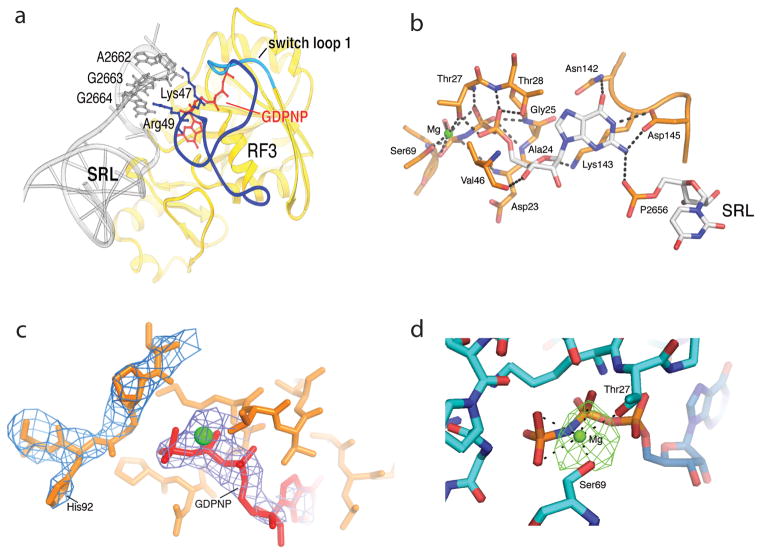
Fig. 5. Ribosome-induced ordering of the GTPase active site of RF3 in the E. coli complex.
(a) Ordering of residues 39–69 of RF3 and re-positioning residues 70–73 (including the switch loop 1 residues 64–72) creates contacts between the G domain of RF3 and the SRL of 23S rRNA. Residues 39–63 are shown in dark blue, the switch loop in cyan, and the rest of RF3 in yellow. (b) An intricate network of H-bonds is formed between the base, ribose and phosphate groups of GDPNP and domain I of RF3, including elements of switch I (residues 64–72), P loop (residues 21–28), NKXD motif (residues142–145) and phosphate 2656 of the SRL of 23S rRNA. A Mg atom (green) is coordinated by Thr27, Ser91 and the β and γ phosphates of GDPNP (see Fig. S9). (c) His92 of RF3 is positioned at a distance of 8 Å from the γ phosphate of GDPNP. The 2Fo-Fc electron density map is shown around His92 and GDPNP, contoured at 1.3 σ and 2.3 σ, respectively. (d) Difference electron density showing Mg bound to GDPNP. An Fo-Fc difference map was calculated using the complete RF3·GDPNP·70S ribosome model, shown here contoured at 4.5 σ. The Mg ion (green) is coordinated with the β- and γ-phosphate oxygens of GDPNP and with the side-chain hydroxyls of Thr27 and Ser 69. From ref. [43].