Abstract
Epigenetic modifications which are defined by DNA methylation, histone modifications and microRNA mediated gene regulation, have been found to be associated with cardiac dysfunction and cardiac regeneration but the mechanisms are unclear. MicroRNA therapies have been proposed for cardiac regeneration and proliferation of stem cells into cardiomyocytes. Cardiovascular disorders are represented by abnormal methylation of CpG islands and drugs that inhibit DNA methyl transferases such as 5-methyl Aza cytidine are under trials. Histone modifications which include acetylation, methylation, phosphorylation, ADP ribosylation, sumoylation and biotinylation are represented within abnormal phenotypes of cardiac hypertrophy, cardiac development and contractility. MicroRNAs have been used efficiently to epigenetically reprogram fibroblasts into cardiomyocytes. MicroRNAs represent themselves as potential biomarkers for early detection of cardiac disorders which are difficult to diagnose and are captured at later stages. Because microRNAs regulate circadian genes, for example a nocturnin gene of circadian clockwork is regulated by mir122, they have profound role in regulating biological clock and this may explain the high cardiovascular risk during the morning time. This review highlights the role of epigenetics which can be helpful in disease management strategies.
Keywords: Epigenetics, stem cells, cardiomyocytes, microRNA, cardiac disorders, biomarkers
Introduction
Epigenetics has emerged as one of the important phenomena behind cardiac disorders and includes small non coding RNAs like microRNAs, DNA methylation and histone modifications (Fig 1). Epigenetics refers to changes in gene expression which are not due to change in the DNA sequence but attributed to chromatin alteration or packaging which changes the accessibility of DNA [1]. Epigenetic changes often are a result of gene environment interactions or surrounding conditions [2] leading to enhanced/decreased expression or silencing of genes e.g. in the case of diabetes, hypertension and obesity. DNA methylation are the most studied epigenetic modifications and mainly involve methylation of CpG islands in the promoter of genes. Diseased states such as cardiovascular disorders are represented by abnormal methylation of the CpG islands consequently leading to modifications in gene expression [3]. Movassagh et al [4] have identified different patterns of DNA methylation in heart failure patients. Recently, homocysteine which is a marker for cardiovascular disease has been shown to play crucial role in epigenetics [5, 6]. Hyoperhomocysteinemia has been reported to be associated with alteration in the DNA mthyltransferase activities involving methionine, folic acid and cystathione B synthase. DNA methylations can be transferred mitoticaly to next generations through cell divisions as they are quite stable. Histone modifications are another mechanisms to altered gene expression via chromatin remodeling by histone acetyltransferases or deacetylases which change the DNA accessibility. The role of histone regulatory proteins in heart disease have been demonstrated by Backs et al [7]. Furthermore, RNA based mechanisms like microRNAs and non-coding RNAs are also involved in altering gene expression of target genes and are studied extensively in cardiac disorders [8–11]. MicroRNAs have also been implicated in stem cell therapies e.g. miR133a has been reported to be involved in differentiation of cardiogenic Mesenchymal stem cells by targeting epidermal growth factors [12]. This review aims to elucidate epigenetic mechanisms underlying cardiac disorders and how these epigenetic mechanisms can be beneficially introduced for cardiac therapies.
Figure 1.
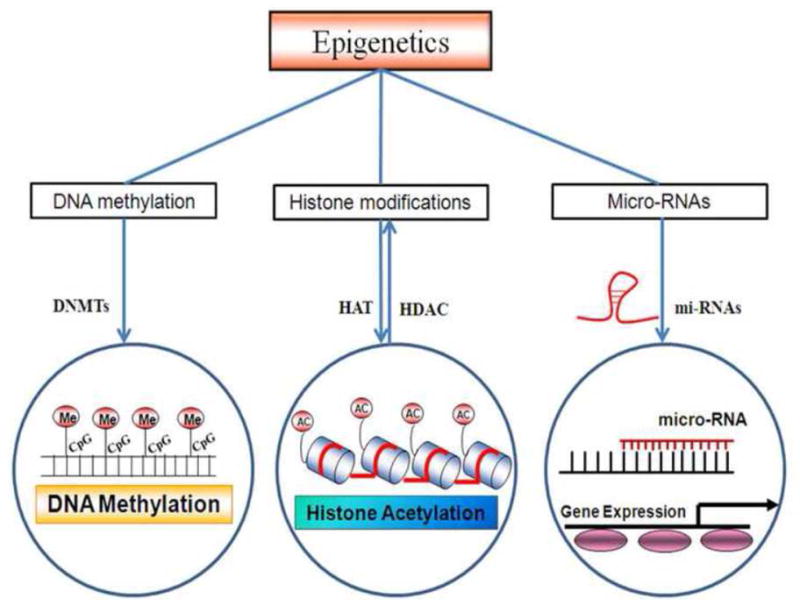
Epigenetic mechanisms and role of DNA methylation, histone modification by acetylation and methylation and post-transcriptional control by microRNA. Mthyl trasferase is involved in gene methylation. HAT and HDAC are involved in histone acetylation and chromatin remodeling. microRNAs are involved in post-transcriptional control of the genes.
Role of epigenetics in Cardiac degeneration
Role of DNA methylation
The pathogenesis of cardiovascular disease is well studied but the role of epigenetics still needs to be explored. The epigenetic modifications in cardimyopathy have been summarized in Table 1. Many studies have demonstrated with help of animal models that DNA methylation plays an important role in atherosclerosis and cardiovascular disease [13]. DNMTs (DNA methyltransferases) and MTHFR (Methylene tetrahydrofolate reductase) represent two important genes in DNA methylation and mice deficient in these genes show hypomethylation of their DNA [13, 14]. Chen et al have shown the formation of aortic fatty streaks in MTHFR deficient mice. In leucocytes of DNMT deficient mice, there is increase in the expression of inflammatory mediators which represent hypomethylation [15]. In peripheral blood leukocytes, there are changes in DNA methylation in ApoE−/− mice, which lead to atherosclerosis promotion and dysregulation of inflammation [14].
Table 1.
Overview of epigenetic modifications: DNA methylation and Histone modifications in cardiomyopathy.
| Epigenetic Modification | Target | Binding domains | Altered Gene expression | Phenotype | References |
|---|---|---|---|---|---|
| DNA methylation | CPG islands | None | Upregulation or Downregulation | Heart failure and altered gene expression of angiogenic factors | [70, 134] |
| Histone modifications | |||||
| Methylation | H3K4me3, H3K4me2, H3K9me3, H3K27me3, H3K79me | PTIP, DOTIL | Upregulation or Downregulation | Fetal cardiac gene activation; angiogenesis and heart failure; dialated cardiomyopathy | [59–61, 143] |
| Demethylation | H3K4me3, H3K4me3, H3K36me3, H3K27me3 | JMJD2A, UTX | Upregulation or Downregulation | Cardiac hypertrophy stimulation; heart malformation and embryo lethality | [59, 62–64] |
| Acetylation | H3K4, K19, H4K5, K8, K12, K16 | CREBP- binding protein (CBP)/p300 | Upregulation | Cardiac hypertrophy regulation | [49, 135, 136] |
| Deacetylation | Histone tails | Class II HDACs (HDAC-4,5,9) | Inhibit the activity of myocyte enhancer factor 2 (MEF2); negative regulation of cardiac hypertrophy | [55, 137] | |
| Phosphorylation | H3 S10/S28/S10, H4Y41 and H2B, HDACs |
Aurora, Rsk2, Msk1, IKKα, PIMI, Akt, CaMK11, JAK2, AMPK, PKD | Upregulation or Downregulation | Mototic activity, cellular proliferation, cardiac hypertrophy regulation; transcriptional activation | [8, 138–142] |
| Ribosylation | Histones and PARP-1, HDACs and brg1 | PARP-1 | Upregulation | Cardiac hypertrophy and heart failure; form a complex with Brg1 and HDACs and increase expression of fetal β-MHC | [52] |
The DNA methylation status is also affected by dietary folate and vitamin levels and providing these dietary supplements to female mice before conception causes higher methylation of CpG islands in the offspring [13]. Due to this the offspring has a characterstic phenotype of brown coat color, insulin resistance, cancers, lengthened life span and reduced susceptibility to obesity. The aortas of ApoE knockout mice represent a decrease in DNA methylation which can be detected at 4 weeks and any histological changes associated with atherosclerosis can be determined [16]. The estrogen receptors α and β show increased methylation at the promoter region in atherosclerotic tissues and the methylation of estrogen receptor increases with age. There is hypermethylation of the HSD11B2 gene, and loss of global methylation of genomic content in blood leukocytes of hypertensive patients [17]. In another report it was shown that in patients with atherosclerotic cardiovascular disease, there is lower DNA methylation in blood leukocytes [18]. Baccarelli et al [19] have shown that incidence and mortality from ischemic heart disease and stroke can be predicted by lower LINE-1 (Long Interspersed Nucleotide Elements) methylation in peripheral blood leukocytes. LINE-1 have been evaluated as surrogate markers for global methylation status [20].
Hypertension which is associated with cardiac degeneration is influenced by global genomic methylation content in the peripheral blood leukocytes [17] of hypertensive patients. In Chinese individuals, elevated Alu methylation status in peripheral blood leucocytes have been related to the prevalence of cardiovascular disease and obesity [21]. Friso et al [22] have linked hypermethylation of the HSD11B2 gene with blood pressure control. Impaired lipid and glucose metabolism which leads to increased cardiovascular risk and diabetes, are represented by hypermethylation of MEG3, IL-10, GNASAS, ABCA1 and hypomethylation of IGF2 and INSIGF genes [23].
Role of microRNAs
MicroRNAs have emerged recently as one of the epigenetic mechanisms underlying cardiovascular diseases (Fig 2, Table 2). In patients with atherosclerotic plaque, the elevated levels of mir127 leads to disruption of endothelium and subsequently, vascular senescence via inhibiting SIRT1 [24]. In animal models and patients with myocardial infarction, mir133b and miR499 have been shown to be upregulated and are potential candidates for biomarkers [8, 25]. Additionally, in patients with coronary artery disease, the level of mir126 and mir145 is decreased profoundly [26]. Downregulation of mir126 indicates inflammation of vessel walls during the development of atherosclerosis by promoting the expression of VCAM-1 [27, 28]. In unstable angina patients, the levels of mir134, mir370 and mir198, were found to be significantly increased which showed increased risk of cardiovascular disease [29]. Sondermeijer et al [30] reported that mir340 and mir624 were significantly increased in patients with cardiovascular diseases.
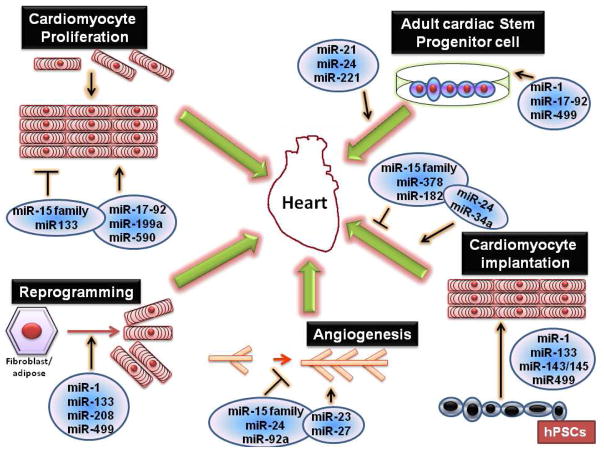
Figure 2.
The role of microRNAs in cardiac regeneration. mir15 family suppresses the proliferation of cardiomyocytes and angiogenesis while mir1, -133 and -499 induces fibroblast/adipocyte reprogramming into cardiomyocytes. Mir1 and -499 also promote the conversion of cardiac stem progenitor cells into cardiomyocytes and they can be injected for cardiac regeneration. hPSC or human pleuripotent stem cells can be converted to cardiomyocytes by the induction of mir1, mir133 and mir499.
Table 2.
miRNAs involved in cardiac degeneration:
| micro-RNA | Fold expression | Site of expression | Methodology | Reference |
|---|---|---|---|---|
| Acute myocardial infarction | ||||
| mir-1 mir-133a mir-208b mir-4993p |
Upregulated 300 times Upregulated 70 times Upregulated 3000 times Upregulated 250 times |
Human Plasma | qRT-PCR | [37] |
|
| ||||
| mir-223 | Downregulated | Human Plasma | qRT-PCR | [33] |
|
| ||||
| mir-21 | Downregulated in infarcted cells Upregulated in border cells |
Rat Myocytes |
qRT-PCR | [32] |
|
| ||||
| mir-122 mir-375 |
Downregulated | Human Plasma | qRT-PCR | [26] |
|
| ||||
| mir-423-5p | Upregulated 3–10 fold | Human Plasma | qRT-PCR | [11] |
|
| ||||
| mir-663-b, -1291, mir-306, -145 | Downregulated Upregulated |
Human PBMCs | Array | [12] |
|
| ||||
| mir-328 | Upregulated in plasma | Human plasma | qRT-PCR | [144] |
|
| ||||
| Coronary Artery Disease | ||||
| mir-17-92 cluster, -126, -145, -155, mir-133, -208a | Downregulated Upregulated |
Human Plasma | Array | [27] |
|
| ||||
| mir-92a, -126, - 133a, -208a, -499 | Upregulated 2–20 fold, ACS vs CAD | Human Plasma | qRT-PCR | [116] |
|
| ||||
| mir-134, -135a, - 147, -198, -370 mir-147 |
Upregulated 3.1–12 fold Downregulated 4 fold |
Human PBMCs | qRT-PCR | [30] |
|
| ||||
| mir-340, mir-624 | Upregulated >1.5 fold | Human Platelets | Array | [31] |
|
| ||||
| Heart failure | ||||
| mir-24, -125b, - 195, -199a, -214 | Upregulated 1.3–3 fold | Human left ventricle | Northern blots | [38] |
|
| ||||
| mir-214 mir-19a/b |
Upregulated 2–2.8 fold Downregulated 2–2.7 fold |
Human left ventricle | Bead based hybridization | [39] |
|
| ||||
| mir-29b, -142-3p mir-107, -125b, - 139, -142-5p, -497 | Upregulated >2 fold Down regulated >2 fold |
Human PBMCs | qRT-PCR | [145] |
|
| ||||
| mir-125, -181b, - 214, -342 | Upregulated | Human left ventricle | Array | [146] |
|
| ||||
| mir-100, -195, | Upregulated | Human heart tissue | Array | [40] |
|
| ||||
| mir-92, -133b mir-21, -129, -212 |
Downregulated Upregulated >1.5 fold |
Human heart tissue | Array | [147] |
The role of miR21 in early phase MI is evident from an animal study, where acute MI created by left vertricular coronary artery ligation leads to decrease in expression of mir21 in the infracted area as compared to the surrounding area [31]. Wang et al [8] found elevated levels of miR208a in CAD patients as compared to healthy subjects, where this microRNA was undetectable. In another study by Corsten et al [32], the levels of miR208a were increased to 90–100 fold. In CAD, miR1 is the most widely studied and plasma miR1 levels are significantly increased in MI patients as compared to healthy controls. This increase was independent of clinical parameters such as age and sex [33, 34]. After the MI event, in a cohort of patients, miR1, miR21, miR133a and miR208 were found to be significantly elevated [35]. Similarly, D’Allessandra et al [25] also showed increase in miR1, miR133a/b, miR499-5p by upto 140 fold in MI patients as a consequence of cardiac injury. They also confirmed these findings in mouse models of coronary artery ligation. In an attempt to determine diagnostic potential of microRNA, Olivieri et al [10] have found increased levels of miR423-5p in congestive heart failure than in MI, while miR499-5p is upregulated in both MI and HF.
MicroRNAs extracted from whole blood have been assessed for diagnostic and prognostic properties, so that even when troponin T and ischemic heart disease biomarkers are negative, the microRNAs can predict MI [11]. miR30c and miR145 showed good correlation with troponin T levels and their expression increased in MI patients. Additionally, mir1291 and mir663b were able to distinguish case-control patients and were highly sensitive. The correlation between indicators of cardiac necrosis (Troponin and cardiac kinase) and circulating microRNA levels have been investigated to find out early biomarkers e.g. mir1 and mir133a/b were detected before troponin 1 increase in patients affected by ST elebvation MI [25]. Similarly, creatine kinase and troponin T levels were significantly correlated with circulating levels of mir208b and mir499 [9, 32, 36]. In elderly patients, mir499-5p is more sensitive marker than troponin T and electrocardiogram and is able to differentiate MI from acute congestive heart failure [10].
In mouse models of cardiac hypertrophy and congestive heart failure, microRNA profiling with microarray revealed five microRNAs (mir24, mir125b, mir195, mir199a, mir214) to be upregulated (Table 2). These findings were also confirmed in patients with heart failure [37]. The authors also showed that over-expression of mir195, correlated with cardiomyocyte disorganization in transgenic mice model, which leads to heart failure. In HF patients with different aetiologies (ischemic, aortic stenosis and dialated cardiomyopathy), the microRNA expression profiling was performed in left ventricular tissue by Ikeda et al [38] and 43 microRNAs were found to be differentially expressed in cardiomyopathy while thirteen microRNAs were specific to aortic stenosis [38]. In two groups of patients affected by idiopathic dialated cardiomyopathy and ischemic cardiomyopathy, the microRNA expression profile specific to each aetiology was determined by Sucharov et al [39]. Several microRNAs common to all failing hearts were also found e.g. mir92 and mir133b were downregulated, while mir195 and mir100 were upregulated [39], which are speculated to prevent changes in β-adrenergic mediated gene expression- an important pathway in heart failure. Matkovich et al [40] performed microarrays and mRNA profiling of myocardial samples and found 28 microRNAs to be upregulated in heart failure. The study suggested that in myocardial response to stress, microRNA can serve as more specific biomarkers [40].
Role of Histone modifications
Histone modifications together with DNA methylation and microRNAs are dynamic processes that lead to chromatin remodeling and modulate gene expression. Histones (H2A, H2B, H3 and H4) which form nucleosome- a basic unit of chromatin, are modified by post translational modifications such as acetylation, methylation, phosphorylation, ADP ribosylation, sumoylation and biotinylation [41, 42] (Fig 1, Table 1). Histone acetylation/deacetylation represents central mechanisms that alter chromatin structure to control gene expression [43–45]. Histone acetylation by histone acetyltransferases (HAT) loosens the histone-DNA interactions, ‘relaxes’ chromatin structure and activates transcription, while deacetylation by histone deacetylases (HDAC) increases histone-DNA interactions, results in chromatin condensation and leads to shut down of transcription machinery. Acetylation of histones by acetyltransferases- p300 or CREBP (cAMP responsive element binding protein), are important in heart development and deletion of either of genes is lethal in the embryo [46]. Ablation of the histone acetyltransferase (HAT) domains in both p300 and CREBP leads to abnormality in the cardiovascular system [47]. P300 acetylates transcription factors such as GATA4 and overexpression of p300 in mouse heart results in depressed cardiac dialation [43, 48, 49].
The expression of eNOS (Nitric Oxide Synthase) in the endothelial cells is important in vascular functioning and is controlled by cell-specific histone modifications [50]. The authors demonstrate that the core promoter of eNOS gene is rich in acetylated H3K9 and H4K12 in the endothelial cells and these histone modifications are functionally relevant for the expression of the eNOS gene. Also the expression of the eNOS gene is remarkably decreased in cardiovascular disease [50]. In a study by Hang et al [51] it was demonstrated that cardiac hyopertrophy and failure is represented by chromatin remodeling and transcriptional reprogramming following myocardial stress. In mouse model, the expression of Brg 1 (Brahma related gene) is increased in hypertrophic cardiomyopathy and repression of this gene decreases hypertrophy and reverses myosin isoform shifts [51].
Histone methylation takes place at arginine or lysine residues at H3K4, H3K9, H3K27, H3K36 and H4K20, with mono-, di- or tri- methylated (H3Kme3) histones [52]. As compared to acetylation, which leads only to active state of chromatin, methylation can lead to active, repressed or ‘poised’ states of chromatin. Many studies have explored the role of histone methylation in cardiac development as well as heart failure and cardiac hyopertrophy [53–56] (Table 1). Stein et al [57], have shown that H3K4 methylation levels are critical for physiological functions in adult murine cardiomyocytes and loss of H3K4 methylation, increases intracellular calcium resulting in increased contractility [57]. The promoter regions of β-MHC (Myosin Heavy Chain) is hypermethylated at H3K4me3 in cardiac hypertrophy, while for α-MHC, the H3K4me3 methylation level is decreased showing both activation and inactivation of genes under pathological stress [57]. Similarly, in heart failure rodents and patients with end stage heart failure, the H3K4me3 and H3K9me3 levels are disturbed [58, 59].
The eNOS (Nitric Oxide Synthase) gene which is important in endothelial function and angiogenesis, is controlled by promoter methylation at H3K27me3 and H3K4me3. To suppress angiogenesis triggered by hypoxia, the ratio of active H3K4me3 to H3K27me3 increases, which leads to increased expression of histone demethylase JMJD3, which suppresses angiogenesis [60]. UTX gene which is a H3K27 demethylase, contains Jumonji C domain and interacts with cardiac specific transcription factors such as GATA4, NKX2.5, TBX5 and SRF [61]. The removal of H3K29me3 is important in heart development and mice deficient in UTX gene suffer severe heart malformation [62]. JMJD2A is another demethylase which belongs to Jumonji C domain- containing family and catalyzes demethylation of H3K9me3 and H3K36me3. The level of methylation of H3K9 and H3K36 is increased in JMD2A deficient mice, however they are resistant to cardiac stress, though they represent normal phenotype under normal conditions [63]. On the other hand overexpression of JMD2 in transgenic mice increases cardiac hypertrophy after pressure overload [63]. JMD2 enhances cardiac hypertrophy by binding of myocardin and SRF transcription factors to FHL1 promoter (Full-and-a-half LIM domains) which leads to reduced H3K9 methylation and thus, proper cardiac functioning after pathological insult [63].
Majority of methylation takes place at the histone tail i.e. H3K9 position, though additional residues such as H3K79, which are located in the globular domain are also subject to methylation. The DOTIL (Disruption of Telomeric silencing protein) catalyzes the methylation of H3K79 and deletion of DOTIL in mice causes dialation of cardiac chambers, reduced contractility and increased lethality [64]. Deletion of DOTIL gene also reduces H3K79 methylation at the dystrophin promoter and decreases the transcription of dystrophin gene which is important for cardiac muscle functioning [59]. Additionally, in patients with idiopathic dialated cardiomyopathy, DOTIL gene is downregulated and H3K9 methylation is impaired which leads to reduced contractility [64].
The importance of histone modifications, have triggered exploration of whole genome studies which includes co-immunioprecipitaiton (ChIP) and ChIP with microarray analysis (ChIP-ChIP). The method of choice is ChIP-followed by massive sequencing which gives information about global histone modifications and gene expression on large scale. Hence studies have revealed that different regions in the gene are associated with different histone modification patterns and different load of gene activity. The active genes clearly exhibit histone acetylation, in the promoter regions while methylation can be detected in either active or repressed genes, hence active and repressive states can co-exist within the same inactive promoters [64–67]. Till date, there are two genome wide studies for hostone modifications in failing heart [68, 69]. One study states that during development of heart failure the genes of calcium signaling and cardiac contractility exhibit differential methylation patterns at H3K4me3 and H3K9me3. The other study, demonstrates that H3K36me3 is enriched in activity transcribed regions of the genome in patients with end stage heart failure.
Apart from the role of epigenetics in cardiovascular diseases such as hypertension, acute MI and congestive heart failure, the relation between these heart diseases and myocardial degeneration remain elusive. Cardiac degeneration is underlined by cell death which is primarily caused by apoptosis, necrosis or autophagic mechanisms. In models of MI, cell death takes place within the area of ischemia over the first 6–24 hrs [70]. In the ischemic zone, cardiomyocytes die by both apoptosis and necrosis. Studies have also shown autophagy associated cell death in myocardial infarction [71–74]. Although in failing hearts, the cell death is defined by apoptosis, necrosis and autophagy, the percentage of these three mechanisms in contributing to cell death varies and depends upon the pathology of the disease [75–77]. The knowledge of cell death and its regulation may be helpful in manipulating death pathways to therapeutic advantages.
Role of epigenetics in cardiac regeneration
Role of microRNAs
Heart development during embryonic stages involves increase in the number of cardiomyocytes for heart enlargement, however, after a short time of birth, cardiomyocytes stop proliferating [78–80]. In adults, cardiac proliferation occurs mainly through hypertrophic enlargement of cardiomyocytes, though the proliferation capacity of cardiomyocytes to proliferate is limited [81, 82] (Fig 2). Hence, the ability of heart to regenerate and repair itself after injury such as myocardial infarction or heart failure is very limited [83–87]. Mollova et al [88] have reported that cardiomyocyte proliferation occurs at the rate of 900 million per year upto one year after birth and it decreases to 70 million per year by the age of 20 [88]. Cardiomyocytes have been associated with altered expression of microRNAs and microRNAs involved in cardiomyocyte proliferation have also been identified [89] (Fig 2, Table 3). Eualalilo et al screened a library of 875 microRNA mimics (http://mirbase.org) and identified 204 microRNAs that increased proliferation in neonatal rat cardiomyocytes and 40 microRNAs that proliferated mouse cardiomyocytes (Table 3). The authors found two microRNAs mir199a-3p and mir590-3p that most efficiently promote cardiomyocyte proliferation in both mouse and rat. Porello et al [90] showed that the regenerative capacity of neonatal heart is more than the adult in mammals and the regenerative potential of neonatal mouse heart after resection of left ventricle is lost after first postnatal week. There is upregulation in the expression of mir15 gene family members with the loss of regeneration potential within first postnatal week.
Table 3.
mi-RNAs involved in cardiac regeneration
| micro-RNA | Functions | Reference |
|---|---|---|
| miR-1 | Increases differentiation of hPSCs into cardiomyocytes and enhances direct reprogramming of human fibroblasts into cardiomyocytes | [89, 91, 92, 148] |
| miR-499 | Overexpression enhances hPSC cardiac differentiation and CPC cardiac differentiation | [89, 92] |
| miR-199a | Expression in cardiomyocytes induces proliferation and facilitates cardiac regeration following myocardial infarction in adult mice | [82] |
| miR-15 family | Inhibition of this microRNA enhances cardiomyocyte survival following ischemia-reperfusion injury in vitro and in vivo, as well as in response to palmitate in vitro; inhibition also facilitates cardiac regeration following myocardial infarction in adult mice | [83, 149–152] |
| miR-199b | In vivo inhibition of miR-199b normalized significantly attenuated cardiac functional impairment, fibrosis and NFAT activity; the target of miR-199b is Dyrk1 | [153] |
| mir-29 | miR-29 downregulation prenvents aortic dialation | [154] |
| miR-21 | Antagomir-21 decreased the development of cardiac fibrosis and improved cardiac function | [155–157] |
| miR-98 | Overexpression of miR-98 reduced AngII mediated fibrosis and amount of apoptotic myocytes | [158] |
| miR-17-92 cluster | Overexpression enhances cardiac progenitor cell proliferation and facilitates cardiac regeneration following myocardial MI in adult mice | [90] |
| miR-23, miR-24 | Overexpression Enhances cardiomyocyte survival following MI n in adult mice and enhances angiogenesis | [159, 160] |
| mir-27 | Enhances angiogenesis | [160] |
| miR-34a | Expression levels gradually increase with age in mice and humans and inihibition enhances cardiomyocyte survival following MI in adult mice | [154] |
| miR-92a, miR-133 | Enhances mesodermal differentiation of hPSCs and enhances direct reprogramming of human fibroblasts into cardiomyocytes; | [91, 121, 148] |
| miR-143/145 | Knockdown prevents cardiogenesis in murine embryonic stem cell differentiation | [161] |
| miR-378 | Inhibition of this microRNA improves cardiomyocyte survival in an in vitro model of hypoxia-reperfusion | [162] |
| miR-590 | Expression of this microRNA in cardiomyocytes induces proliferation and facilitates cardiac regeration following MI in adult mice | [82] |
Senyo et al [91] have shown that mammalian heart regenerates by the division of pre-existing cardiomyocytes after myocardial injury. These findings are in agreement with the studies in zebra fish where cardiac regeneration has been shown to occur through pre-existing cardiomyocytes [92, 93]. Jayawardena et al [94] have shown that fibroblasts can be directly converted into cardiomyocyte lineage by using specific microRNAs (mir1, 133a, 208 and 499). The converted cardiomyocytes possess spontaneous Ca2+ oscillations, mechanical contractions and proteins specific for cardiomyocytes. Several studies have shown that the proliferation and differentiation of cardiomyocytes from human embryonic stem cells [95] or from human progenitor cardiomyocytes [96] is regulated by microRNAs (mir1, 133a and 499). Additionally, mir17-92 cluster has been identified as an important regulator of cardiomyocyte proliferation by deletion/overexpression of this cluster in adult cardiomyocytes [97]. The authors suggest that mir17-92 cluster can become a therapeutic target for heart regeneration and cardiac repair. The delivery of microRNAs specific to target cells is important in modulating cardiomyocyte proliferation as there may be off target inhibition or activation of microRNAs in non-cardiomyocytes. New techniques are required to deliver microRNAs to specific targets and to address potential safety concerns.
Role of stem cell epigenetics
Stem cells have been used efficiently in cardiac regeneration for the past few years; however, the clinical trials with stem cell therapy have not been far successful and have been modest or short lived. The limitations that need to be addressed in order to improve efficiency of stem cell therapy are 1) stem cell homing and survival; 2) tumorogenicity of pleuripotent stem cells; 3) stem cell should be derived from appropriate cell source and; 4) differentiation of stem cells towards cardiomyocyte lineage. Some of these problems have been addressed by the use of microRNAs to modify stem cells and proliferate them into cardiomyocytes [12]. The cardiomyocytes derived from embryonic stem cells (ECS) have been found to have elevated expression of mir1, -208, -133 and -499 during differentiation [95, 96, 98]. Micro RNA-1 overexpression induces cardiac differentiation in mouse and human embryonic stem cells while mir133 opposes cardiomyocyte differentiation and represses cardiac gene markers in ESC [96, 98]. Inhibition of mir499 blocks differentiation of cardiomyocytes in vitro [95, 96, 99] and overexpression of mir499 in ECS and cardiac progenitor cells induces cardiac gene markers. Mir499 has been proposed to act through the repression of SOX6 transcription factor [96].
When embryonic stem cells expressing mir1 were transplanted to mouse heart with infarction, it protected from ischemic injury in the infarction zone [100, 101]. Similarly, in c-kit+ cardiac progenitor cells, overexpression of mir499, enhances cardiomyocyte differentiation and restores myocardial zone after injecting it into rat heart with myocardial infarction [102]. These studies demonstrate that stem cells can be differentiated effectively into cardiomyocytes by using microRNAs in vivo and in vitro to improve stem cell therapies. Apart from the exogenous transplantation of modulated stem cells, microRNAs have been efficiently used to induce the endogenous cardiac progenitor stem cells. For example, mir17 has been overexpressed in the adult c-kit+ progenitor cells in vivo and resulted in the proliferation of cardiac progenitor cells [103]. Alternative to stem cell approach, researchers have also used fibroblast reprogramming into cardiomyocytes (Fig 2), which has an advantage that after myocardial infarction, the human heart provides rich endogenous supply of fibroblasts. Leda et al [104] have demonstrated that by using the transcription factors i.e. GATA4, MEF2c and TBX5, mouse fibroblasts can be programmed into cardiomyocytes in vitro. The use of fibroblasts and their programming into cardiomyocytes in the infracted heart have been demonstrated efficiently by Quian et al and Song et al [105, 106]. Apart from this, viral vectors such as lentivirus have also been used to carry microRNAs to the infacrted heart and the conversion of fibroblasts into cardiomyocytes in situ [94].
Mesenchymal stem cells (MSC) derived from bone marrow have been used for transplantation into infracted myocardium and resulted in enhanced myocardial regeneration, reduced infarct size and improved cardiac function [107–111]. In patients with acute MI and chronic ischemia, clinical studies using MSC have documented safety and feasibility [112–116]. MSC mediate myocardial tissue repair by two processes: 1) transdifferentiation into cardiomyocytes and endothelial cells and; 2) release of cardioprotective factors which are biologically active, proangiogenic and termed as paracrine factors [117–122]. The heart regeneration capacity of MSC can be enhanced by modulating the expression of certain genes and microRNA. Yu et al [123] demonstrated that cardiomyocyte protection is enhanced by the expression of GATA-4 gene in MSC, which leads to improved left ventricular function and smaller infarct size. The MSC transfected with GATA-4 released more growth factors and increased mir221 expression, which is delivered to adjacent cells through microvesicles and helps in cardioprotection. In another study by Cai et al [124] it was demonstrated that mir124 regulated cardiomyocyte differentiation of bone marrow derived MSC by targeting STAT3 mRNA. Similarly, mir16 has been reported to enhance the differentiation of bone marrow derived mesenchymal stem cells into cardiac phenotype [125]. Zhang et al [126] have reported the cardiac differentiation of Bone marrow derived mesenchymal stem cells (BMSC) in rat by overexpressing mir499 which increased the expression of cardiac specific genes such as NKx2.5, GATA-4, MEF2C and cTnl, hence activating the wnt/β-catenin signaling pathway. Wen et al [127] have summarized the role of microRNAs in regulating BMSC for post-MI cardiac repair.
The regeneration potential of BMSC decreases with increasing age due to changes in cellular functions. Ayala-Lugo et al [128] demonstrated that the availability and functionality of bone marrow stem cells depends upon age, by using experimental model of acute and chronic myocardial infarction. The study explained that the age and the time of treatment post-MI, act synergistically to determine the efficacy of bone marrow stem cell treatment of the infracted heart. In another study by Yu et al [129], the regeneration potential of BMSC isolated from different age groups of rhesus monkeys (<5; 8–10; >12yrs) was tested and it was found that BMSC showed significant changes in the microRNA expressions and heat shock proteins, but non-significant changes in histone modifications. Although aging is associated with poor response of BMSC, the angiogenesis in ischemic tissues can be improved by exercise [130, 131]. Cheng et al [132] have shown that in mice of advanced age exercise training can activate ischemia induced neovascularization by reactivating Akt-dependent hypoxia induced factor (HIF)-1α. In an attempt to explain, why vascular regeneration declines with aging, Di Q et al [133] used deferoxamine which is a stabilizer of HIF-1α and demonstrated that age related decline is due to impairment of cross activation between β3 and VEGFR-2 in EPCs which is partially associated with decreased HIF-1α stability.
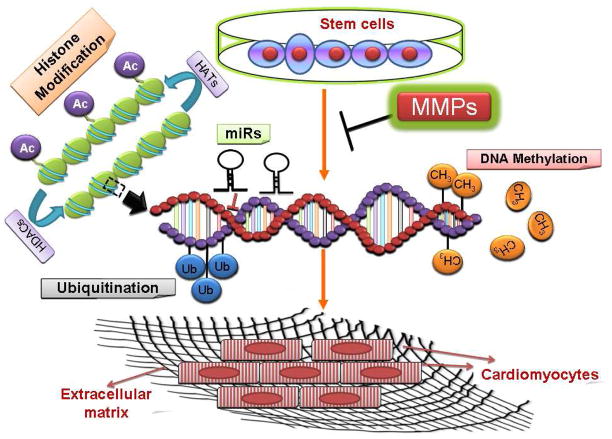
We have shown that ablation of MMP9 leads to the profound conversion of cardiac stem cells into cardiomyocytes [134, 135]. On the basis of our observation and available literature we postulate that microRNAs can lead to the conversion of stem cells to cardiomyocytes through regulating the expression of MMP and DNA methyltransferases (Fig 3). The upregulation of MMP9 gene expression leads to the stiffness of extracellular matrix and consequently fibrosis and affect the differentiation of stem cells into cardiomyocytes.
Figure 3.
Proposed hypothesis for the conversion of stem cells into cardiomyocytes. Based on our previous reports we postulate that microRNAs alter the expression of MMP/TIMP genes which are responsible for matrix degradation and the expression of DNA methyltransferases which consequently lead to altered DNA methylation status. This helps in the conversion of embryonic stem cells to cardiomyocytes.
Epigenetic biomarkers
Micro RNAs have been proposed as biomarkers for the diagnosis of various cardiac diseases, as there is an altered expression of genes and micro RNAs in diseased states and they not only reside in the cells but are abundant in circulating fluids. Disease specific signatures of microRNAs have been identified for coronary artery disease (CAD)[29], heart failure [136] and atherosclerosis [137, 138]. microRNAs can be detected by using conventional techniques such as Nothern Blotting, qRTPCR and advanced techniques such as deep sequencing and genome wide microRNA profiling techniques [139, 140]. Chen et al [141] have shown that circulating microRNAs are quite stable at low and high pH, freeze thaw cycles and boiling which makes them suitable candidates as biomarkers [141]. The stability of microRNAs has been attributed to 1) the formation of protein-microRNA complexes [142, 143]; 2) binding with high density lipoproteins (HDL) and; 3) encapsulation in microvesicles [144, 145] and exosomes [146–148]
Mir423-5p has been shown to be a promising potential candidate in blood samples and cardiomyocytes of patients with heart failure [136] along with mir145, -155, -92a, -17 and -126, in CAD patients [26]. Kuwabara et al [149] have showed that in patients with cardiovascular disease, there is an increased level of mir1 and mir133a in the serum [149]. The altered expression of microRNAs is due to 1) changes in the microRNA production 2) post-transcriptional processing and 3) release of microRNAs from cells. To potentiate the use of microRNA as biomarkers, 20 clinical studies have been performed for circulating microRNAs for MI [36, 96, 150]. The studies show that mir1, -133a/b, -208, -499 [151–153] can be used as potential candidates for biomarker studies in myocardial damage. For vascular wall damage, mir126 [154] mir92a [155] and mir145 [156] have been found to be suitable biomarkers. Mir146, mir155 and mir223 have been found to be associated with leukocytes and platelets and can also be monitored for early stress and myocardial damage [29, 36, 157]. Although microRNAs appear to be very promising as biomarkers still their clinical applications have been delayed due to 1) their laborious isolation and detection procedures as compared to other biomarkers and 2) the time of detection i.e. the time for which they appear in the circulation. However, the combined selection of traditional detection kits and circulating mi-RNAs together, can be used for increasing the specificity and sensitivity of the cardiac diagnostic procedures.
Effect of environment on epigenetics of cardiac disorders
Heart function is affected by environmental factors such as life style, dietary habits, exercise, smoking, alcohol intake and depression. The adverse phenotype faced in later stages of life are due to epigenetic changes, induced by the exposure to harsh conditions in early stages of life [158, 159]. The maternal diet during pregnancy, can dramatically affect the DNA methylation pattern of specific genes which lead to permanent phenotypic changes such as body weight, blood pressure and coat color [160, 161]. The humans who were exposed to Dutch Hunger [23, 162] famine in utero had different DNA methylation patterns as compared to those who were not affected. The risk of cardiac disease is much more in individuals who have prenatal exposure to tobacco smoke. Breton et al [163] have showed altered DNA methylation in LINE-1 (Long Interspersed Elements) and Alu elements in children exposed to smoke in prenatal conditions [163]. The study shows that in utero exposure to tobacco smoke for a long time results in altered DNA methylation. Monozygotic twins gradually become different in their behavior due to epigenetic drift with advancing age caused by different life styles [164]. The pollution created by traffic is associated with increased CVD risk and DNA methylation of LINE-1 and Alu elements [165, 166]. It has been suggested that microRNAs are important regulators of circadian rhythm which can help in the understanding of biological clock [167] and the fact that risk of CVD is more in the early morning than in the late afternoon or late evening further speculates the role of epigenetics.
Future prospects and challenges
In the past few years, microRNAs have emerged as one of the important epigenetic mechanisms behind cardiac dysfunction (myocardial infarction, heart failure, cardiovascular disease and hypertrophy) and regeneration (cardiomyocyte progenitor cells and stem cell proliferation to caridiomyocytes). MicroRNAs have the potential to become pharmacological targets, as dysregulation of a single microRNA can lead to cardiac disorders and their expression can be modulated for stem cell proliferation. This perpetuates the development of therapeutic and diagnostic strategies by regulating the expression of target genes through microRNAs and their detection in body fluids much before the onset of the disease. Understanding the role of histone modifications in differential regulation of genes under diseased conditions can help in mechanistic and disease therapeutics. DNA methylation underlying developmental processes and progression of disease are important phenomena in disease pathologies. However, the translational means of this research are still in their infancy and less efficient. Though, keeping in mind their immense potential, ongoing exploration and refinement of experimental approaches is the need of the hour and good reason to be optimistic.
Abbreviations
- ABCA1
ATP Binding Cassette Transporter A1
- CAD
Coronary Artery Disease
- CREBP
cAMP (Adenosine 3′5′ Cyclic Monophosphate) Response Element-Binding Protein
- CVD
Cardiovascular Disease
- DNMT
DNA methyltransferase
- DOTIL
Disruption of Telomeric silencing protein
- FHL1
Four and a Half LIM domains 1
- GNASAS
Guanine Nucleotide Binding Protein (G Protein), Alpha Stimulating Activity Polypeptide
- HAT
Histone Acetyl transferase
- HDAC
Histone Deacetylase
- HF
Heart Failure
- HSD11B2
Hydroxysteroid (11-beta) dehydrogenase 2
- IL-10
Interleukin 10
- INSIGF
Insulin Induced Gene
- JMJD
Jumonji Domain
- LIM1
Lin11/Isl1/Mec3 transcription factors
- LINE-1
Long Interspersed Neucleotide Elements
- MEG3
Maternally expressed Gene 3
- MI
Myocardial Infarction
- MTHFR
Methylene tetrahydrofolate reductase
- SRF
Serum Response Factor
- TBX5
T Box 5
- UTX
Ubiquitously transcribed tetratricopeptide repeat, X chromosome
- VCAM-1
Vascular Cell Adhesion Molecule 1
Footnotes
Part of this study was supported by NIH grants HL-74185 and HL-108621
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–8. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 2.Dawber TR, Meadors GF, Moore FE., Jr Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41:279–81. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baccarelli A, Rienstra M, Benjamin EJ. Cardiovascular epigenetics: basic concepts and results from animal and human studies. Circ Cardiovasc Genet. 2010;3:567–73. doi: 10.1161/CIRCGENETICS.110.958744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Movassagh M, Choy MK, Goddard M, Bennett MR, Down TA, Foo RS. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PloS one. 2010;5:e8564. doi: 10.1371/journal.pone.0008564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalani A, Kamat PK, Tyagi SC, Tyagi N. Synergy of Homocysteine, MicroRNA, and Epigenetics: A Novel Therapeutic Approach for Stroke. Mol Neurobiol. 2013;48:157–68. doi: 10.1007/s12035-013-8421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123:2145–56. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circulation research. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 8.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. European heart journal. 2010;31:659–66. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 9.Devaux Y, Vausort M, Goretti E, Nazarov PV, Azuaje F, Gilson G, et al. Use of circulating microRNAs to diagnose acute myocardial infarction. Clin Chem. 2012;58:559–67. doi: 10.1373/clinchem.2011.173823. [DOI] [PubMed] [Google Scholar]
- 10.Olivieri F, Antonicelli R, Lorenzi M, D’Alessandra Y, Lazzarini R, Santini G, et al. Diagnostic potential of circulating mir499-5p in elderly patients with acute non ST-elevation myocardial infarction. International journal of cardiology. 2013;167:531–6. doi: 10.1016/j.ijcard.2012.01.075. [DOI] [PubMed] [Google Scholar]
- 11.Meder B, Keller A, Vogel B, Haas J, Sedaghat-Hamedani F, Kayvanpour E, et al. MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res Cardiol. 2011;106:13–23. doi: 10.1007/s00395-010-0123-2. [DOI] [PubMed] [Google Scholar]
- 12.Lee SY, Ham O, Cha MJ, Song BW, Choi E, Kim IK, et al. The promotion of cardiogenic differentiation of hMSCs by targeting epidermal growth factor receptor using microRNA-133a. Biomaterials. 2013;34:92–9. doi: 10.1016/j.biomaterials.2012.09.069. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, Karaplis AC, Ackerman SL, Pogribny IP, Melnyk S, Lussier-Cacan S, et al. Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum Mol Genet. 2001;10:433–43. doi: 10.1093/hmg/10.5.433. [DOI] [PubMed] [Google Scholar]
- 14.Lund G, Andersson L, Lauria M, Lindholm M, Fraga MF, Villar-Garea A, et al. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. The Journal of biological chemistry. 2004;279:29147–54. doi: 10.1074/jbc.M403618200. [DOI] [PubMed] [Google Scholar]
- 15.Makar KW, Wilson CB. DNA methylation is a nonredundant repressor of the Th2 effector program. J Immunol. 2004;173:4402–6. doi: 10.4049/jimmunol.173.7.4402. [DOI] [PubMed] [Google Scholar]
- 16.Turunen MP, Aavik E, Yla-Herttuala S. Epigenetics and atherosclerosis. Biochim Biophys Acta. 2009;1790:886–91. doi: 10.1016/j.bbagen.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Smolarek I, Wyszko E, Barciszewska AM, Nowak S, Gawronska I, Jablecka A, et al. Global DNA methylation changes in blood of patients with essential hypertension. Med Sci Monit. 2010;16:CR149–55. [PubMed] [Google Scholar]
- 18.Castro R, Rivera I, Struys EA, Jansen EE, Ravasco P, Camilo ME, et al. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003;49:1292–6. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- 19.Baccarelli A, Wright R, Bollati V, Litonjua A, Zanobetti A, Tarantini L, et al. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology. 2010;21:819–28. doi: 10.1097/EDE.0b013e3181f20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PloS one. 2010;5:e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friso S, Pizzolo F, Choi SW, Guarini P, Castagna A, Ravagnani V, et al. Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis. 2008;199:323–7. doi: 10.1016/j.atherosclerosis.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 23.Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–53. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menghini R, Casagrande V, Cardellini M, Martelli E, Terrinoni A, Amati F, et al. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation. 2009;120:1524–32. doi: 10.1161/CIRCULATIONAHA.109.864629. [DOI] [PubMed] [Google Scholar]
- 25.D’Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. European heart journal. 2010;31:2765–73. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, et al. Circulating microRNAs in patients with coronary artery disease. Circulation research. 2010;107:677–84. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 27.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 28.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1516–21. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoekstra M, van der Lans CA, Halvorsen B, Gullestad L, Kuiper J, Aukrust P, et al. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochemical and biophysical research communications. 2010;394:792–7. doi: 10.1016/j.bbrc.2010.03.075. [DOI] [PubMed] [Google Scholar]
- 30.Sondermeijer BM, Bakker A, Halliani A, de Ronde MW, Marquart AA, Tijsen AJ, et al. Platelets in patients with premature coronary artery disease exhibit upregulation of miRNA340* and miRNA624*. PloS one. 2011;6:e25946. doi: 10.1371/journal.pone.0025946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, et al. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. The Journal of biological chemistry. 2009;284:29514–25. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010;3:499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 33.Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochemical and biophysical research communications. 2010;391:73–7. doi: 10.1016/j.bbrc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Cheng Y, Tan N, Yang J, Liu X, Cao X, He P, et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119:87–95. doi: 10.1042/CS20090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zile MR, Mehurg SM, Arroyo JE, Stroud RE, DeSantis SM, Spinale FG. Relationship between the temporal profile of plasma microRNA and left ventricular remodeling in patients after myocardial infarction. Circ Cardiovasc Genet. 2011;4:614–9. doi: 10.1161/CIRCGENETICS.111.959841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gidlof O, Andersson P, van der Pals J, Gotberg M, Erlinge D. Cardiospecific microRNA plasma levels correlate with troponin and cardiac function in patients with ST elevation myocardial infarction, are selectively dependent on renal elimination, and can be detected in urine samples. Cardiology. 2011;118:217–26. doi: 10.1159/000328869. [DOI] [PubMed] [Google Scholar]
- 37.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, et al. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–73. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 39.Sucharov C, Bristow MR, Port JD. miRNA expression in the failing human heart: functional correlates. J Mol Cell Cardiol. 2008;45:185–92. doi: 10.1016/j.yjmcc.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, et al. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–71. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 42.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–96. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–52. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 44.Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–8. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 45.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–45. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 46.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–72. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 47.Shikama N, Lutz W, Kretzschmar R, Sauter N, Roth JF, Marino S, et al. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J. 2003;22:5175–85. doi: 10.1093/emboj/cdg502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yanazume T, Hasegawa K, Morimoto T, Kawamura T, Wada H, Matsumori A, et al. Cardiac p300 is involved in myocyte growth with decompensated heart failure. Mol Cell Biol. 2003;23:3593–606. doi: 10.1128/MCB.23.10.3593-3606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyamoto S, Kawamura T, Morimoto T, Ono K, Wada H, Kawase Y, et al. Histone acetyltransferase activity of p300 is required for the promotion of left ventricular remodeling after myocardial infarction in adult mice in vivo. Circulation. 2006;113:679–90. doi: 10.1161/CIRCULATIONAHA.105.585182. [DOI] [PubMed] [Google Scholar]
- 50.Fish JE, Matouk CC, Rachlis A, Lin S, Tai SC, D’Abreo C, et al. The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code. The Journal of biological chemistry. 2005;280:24824–38. doi: 10.1074/jbc.M502115200. [DOI] [PubMed] [Google Scholar]
- 51.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–7. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14967–72. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 54.Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- 55.van Weerd JH, Koshiba-Takeuchi K, Kwon C, Takeuchi JK. Epigenetic factors and cardiac development. Cardiovasc Res. 2011;91:203–11. doi: 10.1093/cvr/cvr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vallaster M, Vallaster CD, Wu SM. Epigenetic mechanisms in cardiac development and disease. Acta Biochim Biophys Sin (Shanghai) 2012;44:92–102. doi: 10.1093/abbs/gmr090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stein AB, Jones TA, Herron TJ, Patel SR, Day SM, Noujaim SF, et al. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. J Clin Invest. 2011;121:2641–50. doi: 10.1172/JCI44641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lorenzen JM, Martino F, Thum T. Epigenetic modifications in cardiovascular disease. Basic Res Cardiol. 2012;107:245. doi: 10.1007/s00395-012-0245-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen AT, Xiao B, Neppl RL, Kallin EM, Li J, Chen T, et al. DOT1L regulates dystrophin expression and is critical for cardiac function. Genes Dev. 2011;25:263–74. doi: 10.1101/gad.2018511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohtani K, Vlachojannis GJ, Koyanagi M, Boeckel JN, Urbich C, Farcas R, et al. Epigenetic regulation of endothelial lineage committed genes in pro-angiogenic hematopoietic and endothelial progenitor cells. Circulation research. 2011;109:1219–29. doi: 10.1161/CIRCRESAHA.111.247304. [DOI] [PubMed] [Google Scholar]
- 61.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 62.Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang QJ, Chen HZ, Wang L, Liu DP, Hill JA, Liu ZP. The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. J Clin Invest. 2011;121:2447–56. doi: 10.1172/JCI46277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 65.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 66.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15782–7. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–80. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaneda R, Takada S, Yamashita Y, Choi YL, Nonaka-Sarukawa M, Soda M, et al. Genome-wide histone methylation profile for heart failure. Genes Cells. 2009;14:69–77. doi: 10.1111/j.1365-2443.2008.01252.x. [DOI] [PubMed] [Google Scholar]
- 69.Movassagh M, Choy MK, Knowles DA, Cordeddu L, Haider S, Down T, et al. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–22. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kajstura J, Cheng W, Reiss K, Clark WA, Sonnenblick EH, Krajewski S, et al. Apoptotic and necrotic myocyte cell deaths are independent contributing variables of infarct size in rats. Laboratory investigation; a journal of technical methods and pathology. 1996;74:86–107. [PubMed] [Google Scholar]
- 71.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circulation research. 2007;100:914–22. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 72.Sciarretta S, Zhai P, Shao D, Maejima Y, Robbins J, Volpe M, et al. Rheb is a critical regulator of autophagy during myocardial ischemia: pathophysiological implications in obesity and metabolic syndrome. Circulation. 2012;125:1134–46. doi: 10.1161/CIRCULATIONAHA.111.078212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhai P, Sciarretta S, Galeotti J, Volpe M, Sadoshima J. Differential roles of GSK-3beta during myocardial ischemia and ischemia/reperfusion. Circulation research. 2011;109:502–11. doi: 10.1161/CIRCRESAHA.111.249532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. The Journal of biological chemistry. 2006;281:29776–87. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 75.Guerra S, Leri A, Wang X, Finato N, Di Loreto C, Beltrami CA, et al. Myocyte death in the failing human heart is gender dependent. Circulation research. 1999;85:856–66. doi: 10.1161/01.res.85.9.856. [DOI] [PubMed] [Google Scholar]
- 76.Olivetti G, Abbi R, Quaini F, Kajstura J, Cheng W, Nitahara JA, et al. Apoptosis in the failing human heart. The New England journal of medicine. 1997;336:1131–41. doi: 10.1056/NEJM199704173361603. [DOI] [PubMed] [Google Scholar]
- 77.Saraste A, Pulkki K, Kallajoki M, Heikkila P, Laine P, Mattila S, et al. Cardiomyocyte apoptosis and progression of heart failure to transplantation. European journal of clinical investigation. 1999;29:380–6. doi: 10.1046/j.1365-2362.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- 78.Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. 2007;87:521–44. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Amerongen MJ, Engel FB. Features of cardiomyocyte proliferation and its potential for cardiac regeneration. Journal of cellular and molecular medicine. 2008;12:2233–44. doi: 10.1111/j.1582-4934.2008.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bicknell KA, Coxon CH, Brooks G. Can the cardiomyocyte cell cycle be reprogrammed? J Mol Cell Cardiol. 2007;42:706–21. doi: 10.1016/j.yjmcc.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 81.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kajstura J, Urbanek K, Perl S, Hosoda T, Zheng H, Ogorek B, et al. Cardiomyogenesis in the adult human heart. Circulation research. 2010;107:305–15. doi: 10.1161/CIRCRESAHA.110.223024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83.Beltrami AP, Urbanek K, Kajstura J, Yan SM, Finato N, Bussani R, et al. Evidence that human cardiac myocytes divide after myocardial infarction. The New England journal of medicine. 2001;344:1750–7. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- 84.Robledo M. Myocardial regeneration in young rats. Am J Pathol. 1956;32:1215–39. [PMC free article] [PubMed] [Google Scholar]
- 85.Nag AC, Carey TR, Cheng M. DNA synthesis in rat heart cells after injury and the regeneration of myocardia. Tissue Cell. 1983;15:597–613. doi: 10.1016/0040-8166(83)90010-1. [DOI] [PubMed] [Google Scholar]
- 86.Kajstura J, Zhang X, Reiss K, Szoke E, Li P, Lagrasta C, et al. Myocyte cellular hyperplasia and myocyte cellular hypertrophy contribute to chronic ventricular remodeling in coronary artery narrowing-induced cardiomyopathy in rats. Circulation research. 1994;74:383–400. doi: 10.1161/01.res.74.3.383. [DOI] [PubMed] [Google Scholar]
- 87.Reiss K, Kajstura J, Capasso JM, Marino TA, Anversa P. Impairment of myocyte contractility following coronary artery narrowing is associated with activation of the myocyte IGF1 autocrine system, enhanced expression of late growth related genes, DNA synthesis, and myocyte nuclear mitotic division in rats. Exp Cell Res. 1993;207:348–60. doi: 10.1006/excr.1993.1202. [DOI] [PubMed] [Google Scholar]
- 88.Mollova M, Bersell K, Walsh S, Savla J, Das LT, Park SY, et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1446–51. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–81. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 90.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–80. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–6. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–5. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–9. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circulation research. 2012;110:1465–73. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilson KD, Hu S, Venkatasubrahmanyam S, Fu JD, Sun N, Abilez OJ, et al. Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for mir499. Circ Cardiovasc Genet. 2010;3:426–35. doi: 10.1161/CIRCGENETICS.109.934281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sluijter JP, van Mil A, van Vliet P, Metz CH, Liu J, Doevendans PA, et al. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol. 2010;30:859–68. doi: 10.1161/ATVBAHA.109.197434. [DOI] [PubMed] [Google Scholar]
- 97.Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, Zhang Z, et al. mir17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circulation research. 2013;112:1557–66. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–29. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fu JD, Rushing SN, Lieu DK, Chan CW, Kong CW, Geng L, et al. Distinct roles of microRNA-1 and -499 in ventricular specification and functional maturation of human embryonic stem cell-derived cardiomyocytes. PloS one. 2011;6:e27417. doi: 10.1371/journal.pone.0027417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Glass C, Singla DK. MicroRNA-1 transfected embryonic stem cells enhance cardiac myocyte differentiation and inhibit apoptosis by modulating the PTEN/Akt pathway in the infarcted heart. American journal of physiology Heart and circulatory physiology. 2011;301:H2038–49. doi: 10.1152/ajpheart.00271.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Glass C, Singla DK. ES cells overexpressing microRNA-1 attenuate apoptosis in the injured myocardium. Mol Cell Biochem. 2011;357:135–41. doi: 10.1007/s11010-011-0883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hosoda T, Zheng H, Cabral-da-Silva M, Sanada F, Ide-Iwata N, Ogorek B, et al. Human cardiac stem cell differentiation is regulated by a mircrine mechanism. Circulation. 2011;123:1287–96. doi: 10.1161/CIRCULATIONAHA.110.982918. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 103.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–88. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–8. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Poynter JA, Herrmann JL, Manukyan MC, Wang Y, Abarbanell AM, Weil BR, et al. Intracoronary mesenchymal stem cells promote postischemic myocardial functional recovery, decrease inflammation, and reduce apoptosis via a signal transducer and activator of transcription 3 mechanism. J Am Coll Surg. 2011;213:253–60. doi: 10.1016/j.jamcollsurg.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 108.Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson SM, Li B, et al. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410:701–5. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- 109.Uemura R, Xu M, Ahmad N, Ashraf M. Bone marrow stem cells prevent left ventricular remodeling of ischemic heart through paracrine signaling. Circulation research. 2006;98:1414–21. doi: 10.1161/01.RES.0000225952.61196.39. [DOI] [PubMed] [Google Scholar]
- 110.Quevedo HC, Hatzistergos KE, Oskouei BN, Feigenbaum GS, Rodriguez JE, Valdes D, et al. Allogeneic mesenchymal stem cells restore cardiac function in chronic ischemic cardiomyopathy via trilineage differentiating capacity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14022–7. doi: 10.1073/pnas.0903201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Angoulvant D, Ivanes F, Ferrera R, Matthews PG, Nataf S, Ovize M. Mesenchymal stem cell conditioned media attenuates in vitro and ex vivo myocardial reperfusion injury. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation. 2011;30:95–102. doi: 10.1016/j.healun.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 112.Boonbaichaiyapruck S, Pienvichit P, Limpijarnkij T, Rerkpattanapipat P, Pongpatananurak A, Saelee R, et al. Transcoronary infusion of bone marrow derived multipotent stem cells to preserve left ventricular geometry and function after myocardial infarction. Clinical cardiology. 2010;33:E10–5. doi: 10.1002/clc.20545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wollert KC, Meyer GP, Lotz J, Ringes-Lichtenberg S, Lippolt P, Breidenbach C, et al. Intracoronary autologous bone-marrow cell transfer after myocardial infarction: the BOOST randomised controlled clinical trial. Lancet. 2004;364:141–8. doi: 10.1016/S0140-6736(04)16626-9. [DOI] [PubMed] [Google Scholar]
- 114.Assmus B, Rolf A, Erbs S, Elsasser A, Haberbosch W, Hambrecht R, et al. Clinical outcome 2 years after intracoronary administration of bone marrow-derived progenitor cells in acute myocardial infarction. Circulation Heart failure. 2010;3:89–96. doi: 10.1161/CIRCHEARTFAILURE.108.843243. [DOI] [PubMed] [Google Scholar]
- 115.Yousef M, Schannwell CM, Kostering M, Zeus T, Brehm M, Strauer BE. The BALANCE Study: clinical benefit and long-term outcome after intracoronary autologous bone marrow cell transplantation in patients with acute myocardial infarction. Journal of the American College of Cardiology. 2009;53:2262–9. doi: 10.1016/j.jacc.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 116.Meyer GP, Wollert KC, Lotz J, Pirr J, Rager U, Lippolt P, et al. Intracoronary bone marrow cell transfer after myocardial infarction: 5-year follow-up from the randomized-controlled BOOST trial. European heart journal. 2009;30:2978–84. doi: 10.1093/eurheartj/ehp374. [DOI] [PubMed] [Google Scholar]
- 117.Boomsma RA, Geenen DL. Mesenchymal stem cells secrete multiple cytokines that promote angiogenesis and have contrasting effects on chemotaxis and apoptosis. PloS one. 2012;7:e35685. doi: 10.1371/journal.pone.0035685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zeng L, Hu Q, Wang X, Mansoor A, Lee J, Feygin J, et al. Bioenergetic and functional consequences of bone marrow-derived multipotent progenitor cell transplantation in hearts with postinfarction left ventricular remodeling. Circulation. 2007;115:1866–75. doi: 10.1161/CIRCULATIONAHA.106.659730. [DOI] [PubMed] [Google Scholar]
- 119.Burchfield JS, Dimmeler S. Role of paracrine factors in stem and progenitor cell mediated cardiac repair and tissue fibrosis. Fibrogenesis & tissue repair. 2008;1:4. doi: 10.1186/1755-1536-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gnecchi M, He H, Noiseux N, Liang OD, Zhang L, Morello F, et al. Evidence supporting paracrine hypothesis for Akt-modified mesenchymal stem cell-mediated cardiac protection and functional improvement. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20:661–9. doi: 10.1096/fj.05-5211com. [DOI] [PubMed] [Google Scholar]
- 121.Shabbir A, Zisa D, Suzuki G, Lee T. Heart failure therapy mediated by the trophic activities of bone marrow mesenchymal stem cells: a noninvasive therapeutic regimen. American journal of physiology Heart and circulatory physiology. 2009;296:H1888–97. doi: 10.1152/ajpheart.00186.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nguyen BK, Maltais S, Perrault LP, Tanguay JF, Tardif JC, Stevens LM, et al. Improved function and myocardial repair of infarcted heart by intracoronary injection of mesenchymal stem cell-derived growth factors. Journal of cardiovascular translational research. 2010;3:547–58. doi: 10.1007/s12265-010-9171-0. [DOI] [PubMed] [Google Scholar]
- 123.Yu B, Gong M, Wang Y, Millard RW, Pasha Z, Yang Y, et al. Cardiomyocyte protection by GATA-4 gene engineered mesenchymal stem cells is partially mediated by translocation of mir221 in microvesicles. PloS one. 2013;8:e73304. doi: 10.1371/journal.pone.0073304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cai B, Li J, Wang J, Luo X, Ai J, Liu Y, et al. microRNA-124 regulates cardiomyocyte differentiation of bone marrow-derived mesenchymal stem cells via targeting STAT3 signaling. Stem cells. 2012;30:1746–55. doi: 10.1002/stem.1154. [DOI] [PubMed] [Google Scholar]
- 125.Liu JL, Jiang L, Lin QX, Deng CY, Mai LP, Zhu JN, et al. MicroRNA 16 enhances differentiation of human bone marrow mesenchymal stem cells in a cardiac niche toward myogenic phenotypes in vitro. Life sciences. 2012;90:1020–6. doi: 10.1016/j.lfs.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 126.Zhang LL, Liu JJ, Liu F, Liu WH, Wang YS, Zhu B, et al. Mir499 induces cardiac differentiation of rat mesenchymal stem cells through wnt/beta-catenin signaling pathway. Biochemical and biophysical research communications. 2012;420:875–81. doi: 10.1016/j.bbrc.2012.03.092. [DOI] [PubMed] [Google Scholar]
- 127.Wen Z, Zheng S, Zhou C, Yuan W, Wang J, Wang T. Bone marrow mesenchymal stem cells for post-myocardial infarction cardiac repair: microRNAs as novel regulators. Journal of cellular and molecular medicine. 2012;16:657–71. doi: 10.1111/j.1582-4934.2011.01471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ayala-Lugo A, Tavares AM, Paz AH, Alegretti A, Miquelito L, Bock H, et al. Age-dependent availability and functionality of bone marrow stem cells in an experimental model of acute and chronic myocardial infarction. Cell transplantation. 2011;20:407–19. doi: 10.3727/096368909X519283. [DOI] [PubMed] [Google Scholar]
- 129.Yu JM, Wu X, Gimble JM, Guan X, Freitas MA, Bunnell BA. Age-related changes in mesenchymal stem cells derived from rhesus macaque bone marrow. Aging cell. 2011;10:66–79. doi: 10.1111/j.1474-9726.2010.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jensen L, Bangsbo J, Hellsten Y. Effect of high intensity training on capillarization and presence of angiogenic factors in human skeletal muscle. The Journal of physiology. 2004;557:571–82. doi: 10.1113/jphysiol.2003.057711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ryan NA, Zwetsloot KA, Westerkamp LM, Hickner RC, Pofahl WE, Gavin TP. Lower skeletal muscle capillarization and VEGF expression in aged vs. young men. Journal of applied physiology. 2006;100:178–85. doi: 10.1152/japplphysiol.00827.2005. [DOI] [PubMed] [Google Scholar]
- 132.Cheng XW, Kuzuya M, Kim W, Song H, Hu L, Inoue A, et al. Exercise training stimulates ischemia-induced neovascularization via phosphatidylinositol 3-kinase/Akt-dependent hypoxia-induced factor-1 alpha reactivation in mice of advanced age. Circulation. 2010;122:707–16. doi: 10.1161/CIRCULATIONAHA.109.909218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Di Q, Cheng Z, Kim W, Liu Z, Song H, Li X, et al. Impaired cross-activation of beta3 integrin and VEGFR-2 on endothelial progenitor cells with aging decreases angiogenesis in response to hypoxia. International journal of cardiology. 2013;168:2167–76. doi: 10.1016/j.ijcard.2013.01.240. [DOI] [PubMed] [Google Scholar]
- 134.Mishra PK, Chavali V, Metreveli N, Tyagi SC. Ablation of MMP9 induces survival and differentiation of cardiac stem cells into cardiomyocytes in the heart of diabetics: a role of extracellular matrix. Can J Physiol Pharmacol. 2012;90:353–60. doi: 10.1139/y11-131. [DOI] [PubMed] [Google Scholar]
- 135.Mishra PK, Kuypers NJ, Singh SR, Leiberh ND, Chavali V, Tyagi SC. Cardiac Stem Cell Niche, MMP9, and Culture and Differentiation of Embryonic Stem Cells. Methods Mol Biol. 2013;1035:153–63. doi: 10.1007/978-1-62703-508-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, et al. MiR423-5p as a circulating biomarker for heart failure. Circulation research. 2010;106:1035–9. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 137.Madrigal-Matute J, Rotllan N, Aranda JF, Fernandez-Hernando C. MicroRNAs and atherosclerosis. Curr Atheroscler Rep. 2013;15:322. doi: 10.1007/s11883-013-0322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. Mir33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–3. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 140.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9740–4. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 142.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–59. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Collino F, Deregibus MC, Bruno S, Sterpone L, Aghemo G, Viltono L, et al. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PloS one. 2010;5:e11803. doi: 10.1371/journal.pone.0011803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.El-Hefnawy T, Raja S, Kelly L, Bigbee WL, Kirkwood JM, Luketich JD, et al. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin Chem. 2004;50:564–73. doi: 10.1373/clinchem.2003.028506. [DOI] [PubMed] [Google Scholar]
- 146.Stoorvogel W. Functional transfer of microRNA by exosomes. Blood. 2012;119:646–8. doi: 10.1182/blood-2011-11-389478. [DOI] [PubMed] [Google Scholar]
- 147.Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–8. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 148.Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kuwabara Y, Ono K, Horie T, Nishi H, Nagao K, Kinoshita M, et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet. 2011;4:446–54. doi: 10.1161/CIRCGENETICS.110.958975. [DOI] [PubMed] [Google Scholar]
- 150.De Rosa S, Fichtlscherer S, Lehmann R, Assmus B, Dimmeler S, Zeiher AM. Transcoronary concentration gradients of circulating microRNAs. Circulation. 2011;124:1936–44. doi: 10.1161/CIRCULATIONAHA.111.037572. [DOI] [PubMed] [Google Scholar]
- 151.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kloosterman WP, Steiner FA, Berezikov E, de Bruijn E, van de Belt J, Verheul M, et al. Cloning and expression of new microRNAs from zebrafish. Nucleic Acids Res. 2006;34:2558–69. doi: 10.1093/nar/gkl278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–73. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, et al. The endothelial-specific microRNA mir126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–71. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–3. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 156.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. mir145 and mir143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–10. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Konstandin MH, Aksoy H, Wabnitz GH, Volz C, Erbel C, Kirchgessner H, et al. Beta2-integrin activation on T cell subsets is an independent prognostic factor in unstable angina pectoris. Basic Res Cardiol. 2009;104:341–51. doi: 10.1007/s00395-008-0770-8. [DOI] [PubMed] [Google Scholar]
- 158.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. The New England journal of medicine. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–88. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 160.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bogdarina I, Welham S, King PJ, Burns SP, Clark AJ. Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circulation research. 2007;100:520–6. doi: 10.1161/01.RES.0000258855.60637.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–7. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–8. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 165.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–8. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hoffmann B, Moebus S, Dragano N, Mohlenkamp S, Memmesheimer M, Erbel R, et al. Residential traffic exposure and coronary heart disease: results from the Heinz Nixdorf Recall Study. Biomarkers. 2009;14 (Suppl 1):74–8. doi: 10.1080/13547500902965096. [DOI] [PubMed] [Google Scholar]
- 167.Pegoraro M, Tauber E. The role of microRNAs (miRNA) in circadian rhythmicity. J Genet. 2008;87:505–11. doi: 10.1007/s12041-008-0073-8. [DOI] [PubMed] [Google Scholar]
- 168.Lee SH, Wolf PL, Escudero R, Deutsch R, Jamieson SW, Thistlethwaite PA. Early expression of angiogenesis factors in acute myocardial ischemia and infarction. The New England journal of medicine. 2000;342:626–33. doi: 10.1056/NEJM200003023420904. [DOI] [PubMed] [Google Scholar]
- 169.Gusterson RJ, Jazrawi E, Adcock IM, Latchman DS. The transcriptional co-activators CREB-binding protein (CBP) and p300 play a critical role in cardiac hypertrophy that is dependent on their histone acetyltransferase activity. The Journal of biological chemistry. 2003;278:6838–47. doi: 10.1074/jbc.M211762200. [DOI] [PubMed] [Google Scholar]
- 170.Mathiyalagan P, Chang L, Du XJ, El-Osta A. Cardiac ventricular chambers are epigenetically distinguishable. Cell Cycle. 2010;9:612–7. doi: 10.4161/cc.9.3.10612. [DOI] [PubMed] [Google Scholar]
- 171.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Baek SH. When signaling kinases meet histones and histone modifiers in the nucleus. Mol Cell. 2011;42:274–84. doi: 10.1016/j.molcel.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 173.Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–22. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–5. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Awad S, Kunhi M, Little GH, Bai Y, An W, Bers D, et al. Nuclear CaMKII enhances histone H3 phosphorylation and remodels chromatin during cardiac hypertrophy. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt500. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 176.Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, et al. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24:8374–85. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pandya K, Kohro T, Mimura I, Kobayashi M, Wada Y, Kodama T, et al. Distribution of histone3 lysine 4 trimethylation at T3-responsive loci in the heart during reversible changes in gene expression. Gene Expr. 2012;15:183–98. doi: 10.3727/105221612x13372578119698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang R, Li N, Zhang Y, Ran Y, Pu J. Circulating microRNAs are promising novel biomarkers of acute myocardial infarction. Intern Med. 2011;50:1789–95. doi: 10.2169/internalmedicine.50.5129. [DOI] [PubMed] [Google Scholar]
- 179.Voellenkle C, van Rooij J, Cappuzzello C, Greco S, Arcelli D, Di Vito L, et al. MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genomics. 2010;42:420–6. doi: 10.1152/physiolgenomics.00211.2009. [DOI] [PubMed] [Google Scholar]
- 180.Naga Prasad SV, Duan ZH, Gupta MK, Surampudi VS, Volinia S, Calin GA, et al. Unique microRNA profile in end-stage heart failure indicates alterations in specific cardiovascular signaling networks. The Journal of biological chemistry. 2009;284:27487–99. doi: 10.1074/jbc.M109.036541. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 181.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–67. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 182.Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5588–93. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, et al. Regulation of neonatal and adult mammalian heart regeneration by the mir15 family. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:187–92. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhu H, Yang Y, Wang Y, Li J, Schiller PW, Peng T. MicroRNA-195 promotes palmitate-induced apoptosis in cardiomyocytes by down-regulating Sirt1. Cardiovasc Res. 2011;92:75–84. doi: 10.1093/cvr/cvr145. [DOI] [PubMed] [Google Scholar]
- 185.Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, Lynch JM, et al. Inhibition of mir15 protects against cardiac ischemic injury. Circulation research. 2012;110:71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Spinetti G, Fortunato O, Caporali A, Shantikumar S, Marchetti M, Meloni M, et al. MicroRNA-15a and microRNA-16 impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circulation research. 2013;112:335–46. doi: 10.1161/CIRCRESAHA.111.300418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.da Costa Martins PA, Salic K, Gladka MM, Armand AS, Leptidis S, el Azzouzi H, et al. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat Cell Biol. 2010;12:1220–7. doi: 10.1038/ncb2126. [DOI] [PubMed] [Google Scholar]
- 188.Boon RA, Seeger T, Heydt S, Fischer A, Hergenreider E, Horrevoets AJ, et al. MicroRNA-29 in aortic dilation: implications for aneurysm formation. Circulation research. 2011;109:1115–9. doi: 10.1161/CIRCRESAHA.111.255737. [DOI] [PubMed] [Google Scholar]
- 189.Thum T, Catalucci D, Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc Res. 2008;79:562–70. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 190.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–4. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 191.Thum T, Chau N, Bhat B, Gupta SK, Linsley PS, Bauersachs J, et al. Comparison of different mir21 inhibitor chemistries in a cardiac disease model. J Clin Invest. 2011;121:461–2. doi: 10.1172/JCI45938. author reply 2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yang Y, Ago T, Zhai P, Abdellatif M, Sadoshima J. Thioredoxin 1 negatively regulates angiotensin II-induced cardiac hypertrophy through upregulation of mir98/let-7. Circulation research. 2011;108:305–13. doi: 10.1161/CIRCRESAHA.110.228437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fiedler J, Jazbutyte V, Kirchmaier BC, Gupta SK, Lorenzen J, Hartmann D, et al. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation. 2011;124:720–30. doi: 10.1161/CIRCULATIONAHA.111.039008. [DOI] [PubMed] [Google Scholar]
- 194.Zhou Q, Gallagher R, Ufret-Vincenty R, Li X, Olson EN, Wang S. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8287–92. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Knezevic I, Patel A, Sundaresan NR, Gupta MP, Solaro RJ, Nagalingam RS, et al. A novel cardiomyocyte-enriched microRNA, mir378, targets insulin-like growth factor 1 receptor: implications in postnatal cardiac remodeling and cell survival. The Journal of biological chemistry. 2012;287:12913–26. doi: 10.1074/jbc.M111.331751. [DOI] [PMC free article] [PubMed] [Google Scholar]