Figure 1.
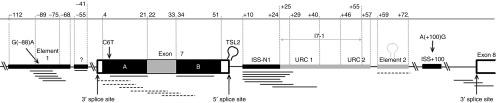
Positive and negative splicing regulatory elements (SREs) modulating SMN2 exon 7 splicing. SMN2 gene depicted from part of intron 6 to partial exon 8 (not drawn to scale). Positive SREs which promotes exon 7 inclusion are shaded in gray whereas negative SREs which inhibit exon 7 inclusion are shaded in black. Published antisense oligonucleotide (AON) target sites are indicated as either full (augmented exon 7 retention) or broken (induced exon 7 skipping) lines below the gene; Element 1,25,42 putative ISS at intron 6,43 exonic splicing silencer (ESS) A,12 exonic splicing enhancer (ESE) at exon 7,12,13 ESS B,12 ISS-N1,13,19,20,21,22,23,24 URC1,19 Element 2,18 and 3′ splice site of exon 8.11,14,44 The relative positions of the polymorphisms are indicated. Numbering of nucleotide position is relative from the first nucleotide: at 3′ of exon 7 in both intron 6 and exon 7, and at 3′ of intron 7 in intron 7 (“+” is appended to differentiate it from exonic sequence). Positive SREs: The exonic positive SRE, which is contiguously flanked by two splicing silencer elements, has been reported to be associated with several serine-rich (SR) or SR-like proteins including Tra2β1, SRp30c, RBMY, and hnRNP-G.12,27,28,29 Correspondingly, antisense oligonucleotides (AONs) bound to this element (depicted as dashed lines) further enhanced exon 7 exclusion in SMN2 transcripts. On the other hand, the three intronic positive SREs are clustered together. Two U-rich clusters (URC1 and URC2) have been reported to be TIA1 binding sites which recruits U1 snRNP needed for splicing.26 Both URC1 and URC2 motifs overlap with I7-1, a 31 nt-segment reported to promote exon 7 inclusion.31 Element 2 was found to be critical for exon 7 inclusion where its stem-loop RNA structure is required to recruit an unidentified splicing protein.18 Indeed, an AON bound to Element 2 promoted exon 7 skipping. Negative SREs: ESSs A and B sandwich the lone positive SRE in exon 7.12 Notably, the C6T transition that occurred within ESS A augments hnRNP A1 binding.4 Element 1, located at intron 6 that encompassed the G(-88)A transition, was reported to be binding sites for hnRNP1 and FUSEBP.25,42 A putative negative SRE was also reported at intron 6 downstream of Element 1.13 Lastly, the remaining two negative SREs, ISS-N1 and ISS+100, flanked all the three intronic positive SREs at intron 7. In fact, ISS-N1 consisted of two weak hnRNAP A1/A2 motifs19 that cooperate to make up a strong splicing inhibitory effect.13 The A(+100)G transition generated a silencer motif at ISS+100 and was reported to have high affinity towards hnRNP A1.33
