Abstract
Stochastic switching is an example of phenotypic bet hedging, where offspring can express a phenotype different from that of their parents. Phenotypic switching is well documented in viruses, yeast, and bacteria and has been extensively studied when the selection pressures vary through time. However, there has been little work on the evolution of phenotypic switching under both spatially and temporally fluctuating selection pressures. Here we use a population genetic model to explore the interaction of temporal and spatial variation in determining the evolutionary dynamics of phenotypic switching. We find that the stable switching rate is mainly determined by the rate of environmental change and the migration rate. This stable rate is also a decreasing function of the recombination rate, although this is a weaker effect than those of either the period of environmental change or the migration rate. This study highlights the interplay of spatial and temporal environmental variability, offering new insights into how migration can influence the evolution of phenotypic switching rates, mutation rates, or other sources of phenotypic variation.
Keywords: stochastic switching, stochastic gene expression variation, evolution in fluctuating environments, metapopulation and migration
GENETICALLY identical cells can show significant cell-to-cell variability in gene expression and other phenotypic characteristics. This variation in gene expression can be due to fluctuations in levels of methylation of CpG sites, in mRNA transcription, or in protein translation and may cause shifts among different regulatory states that result in bi- or multistability (Smits et al. 2006), which have also been viewed as an epigenetic switch (Lim and van Oudenaarden 2007). Moreover, these states are often heritable between generations, perhaps due to epigenetic inheritance. Examples include the lactose utilization network in Escherichia coli, where single cells can stochastically switch between two different states (Mettetal et al. 2006), or the galactose utilization network in yeast that displays bimodal patterns in the expression of GAL genes (Acar et al. 2005; Kaufmann et al. 2007).
In stochastic switching, individual cells can randomly switch between different phenotypes, which may be inherited. The mode of inheritance in such cases can depart from straightforward Mendelism (Bonduriansky and Day 2009; Danchin and Wagner 2010). In particular, there can be interactions between inherited environmental conditions and epigenetic effects (see, e.g., Furrow et al. 2011) that contribute to the statistical heritability of phenotypes.
Why do these genetically identical populations exhibit such switching behaviors? Stochastic switching is often interpreted as a bet-hedging strategy (Starrfelt and Kokko 2012) that could confer a fitness advantage in volatile environments (Thattai and van Oudenaarden 2004; Kussell and Leibler 2005). Switching increases the phenotypic diversity of the population within one generation, thereby increasing the chance of well-adapted offspring in a future environment. Studies have shown that stochastic changes can be advantageous even in comparison to environmental sensing when the switching rate is closely tuned to the rate at which the environment fluctuates (Acar et al. 2008). In this framework, the trade-off between the costs of maladaptation and the benefits of phenotypic variability becomes the object of mathematical analysis (Leigh 1970; Ishii et al. 1989; Thattai and van Oudenaarden 2004; Kussell and Leibler 2005; Leibler and Kussell 2010). It is important to note that bet hedging (increasing phenotypic variance within a generation) as a response to environmental volatility is one way to decrease the variance in fitness between generations and thus increase the geometric mean fitness of the genotype (Carja et al. 2013).
Population genetic approaches to stochastic phenotypic switching have generally been couched in terms of the dynamics of the mutation rate in a fluctuating environment. Mutation, in this context, refers to mutation between different allelic states or epiallelic states that are the cause of the phenotypic variability, and the question boils down to finding the optimal mutation rate as a function of the way the environment changes. Ishii et al. (1989) analyzed one of the earliest models of this kind in which alleles A and a were assigned fitnesses 1 + s(t) and 1 − s(t), respectively, at time t, with s(t) allowed to fluctuate through time with mean zero. A second locus, with alleles B and b, controlled the rate of mutation between A and a. Ishii et al. (1989) found that the evolutionarily stable mutation rate maximized the long-term geometric mean fitness of the population. They suggested that under a wide array of conditions the uninvadable mutation rate was 1/n if the selection cycled with period 2n.
Other studies of symmetric periodic selection models, without the mutation modifying gene as the cause of the switching, also suggested that the fitness was optimized at a mutation rate of 1/n (Lachmann and Jablonka 1996). One important theoretical implication of these results is that if the mutation rate is very low, the population can be invaded by a mutator that increases this rate (up to 1/n). This violates the mutation reduction principle of Feldman and Liberman (1986), which holds in constant environments. When the selection regime does not fluctuate, the reduction principle, that zero is the uninvadable mutation rate, recombination rate (Liberman and Feldman 1986), and migration rate (Balkau and Feldman 1973; Liberman and Feldman 1989), is quite general. However, in fluctuating environments, the reduction principle does not generally hold.
Recent analyses by Salathé et al. (2009) and Liberman et al. (2011) of switching and mutation modification in fluctuating environments have produced results that are much more complicated than originally derived by Ishii et al. (1989). If the time spent in each of two environments is a random variable, then even if the fitnesses in the two environments are symmetric, the stable switching rate can be very different from , where is the mean time in each environment. In addition, if the fitnesses of allele A relative to allele a in one environment and of allele a to allele A in the second environment are not very similar, then a switching–reducing modifier allele will succeed, with the result that the switching rate ultimately evolves toward zero.
In the case of symmetric selection with cycles of period 2, Liberman et al. (2011) showed that for any recombination rate between the switching–modifying gene and the gene under direct selection, higher switching rates are always favored, and the mean fitness at equilibrium is an increasing function of the switching rate; the opposite is true in a constant environment. Liberman et al. (2011) also proved that with period 4 and no recombination, the stable switching rate was 1/2. Furthermore, when there is no recombination, the critical points of the mean fitness with respect to the switching rate are the same switching rates that cannot be invaded. With asymmetric fitnesses, mathematical results have not been obtained, but numerical analysis, with and without recombination, has demonstrated that large asymmetries actually lead to a reduction of the switching rate (Salathé et al. 2009).
The regimes in which the benefits outweigh the costs of switching have been extensively studied both theoretically and experimentally when there is temporal heterogeneity in the selection pressures. However, organisms experience environmental heterogeneity through both time and space. The change in the selection pressure for an organism’s offspring can therefore arise either from a temporal change in the fitness function or from migration to a different deme. Although the evolution of phenotypic switching has been extensively studied when the selection pressures vary temporally, there have been few studies of the evolution of stochastic switching in the case of a subdivided population, when selection varies spatially. We might expect differences in the evolutionary dynamics because in a spatially heterogeneous environment, only the migrants experience the change in the selection pressures, while in temporally varying environments the change in selection pressure is experienced by the entire population. Studies of the evolution of phenotypic variability when there is both spatial and temporal heterogeneity include that of Arnoldini et al. (2012), who investigated the evolution of sensed switching, finding that the relative balance of sensed and stochastic switching depended on the accuracy of the environmental stress signal. Moran (1992) explored the evolution of noninherited phenotypic variability and argued that the optimal amount of variability is zero.
Our goal here is to study the evolution of switching rates in the presence of both spatial and temporal heterogeneity in selection pressures. Do the inherent differences between temporal and spatial change in the selection regime result in different evolutionary conditions under which stochastic switching evolves? We find that the evolutionary dynamics of the system are mainly governed by the rate of environmental change. For large enough periods in the environmental fluctuations, the migration rate in the system also has strong effects on the evolution of stochastic switching: the uninvadable switching rate is on the order of 1/n minus the migration rate, where selection cycles with a period 2n, and zero when the migration rate is >1/n. This stable switching rate is also a decreasing function of the recombination rate in the system, although this effect is weaker than those of the environmental change and migration. The evolution of stochastic switching is therefore determined by a complex interplay between all the forces that contribute to the variability under periodic change in selection pressures.
Modeling Stochastic Switching
Consider an infinite, haploid population, divided into two demes, Ex and Ey, each with its own selection regime. Every individual in this population is defined by two biallelic loci: the major A/a locus controls the phenotype and fitness of the individual, while the modifier B/b locus is selectively neutral and controls the switching rate between these two phenotypic states. Alleles B and b determine switching rates μB and μb, respectively, and our main interest is in finding the evolutionarily stable switching rate between the two phenotypes A and a, namely, a switching rate that cannot be invaded by mutants that produce different rates of switching. Toward that goal, we use an explicit population genetic model to track the allele frequencies at the modifier gene and ask under what conditions phenotypic switching is adaptive and what the role of migration is in the evolutionary dynamics of the modifier locus.
Biological interpretation of the model
Even though the model is presented from the perspective of genetically controlled phenotypic switching (examples of which can be found in Smits et al. 2006; Veening et al. 2008; Beaumont et al. 2009; Rainey et al. 2011) between two bistable states or phenotypes (similar to Thattai and van Oudenaarden 2004; Salathé et al. 2009; Gaál et al. 2010; Liberman et al. 2011), it is also general enough to be applicable to a wider range of evolutionary questions. A possible mechanism for switching is epigenetic control of gene expression through variation in levels of methylation or chromatin formation. Therefore, the B/b modifier locus can be interpreted as an epigenetic locus able to influence the transition between the two phenotypes or the levels of expression of alleles A and a. Examples of such epigenetic loci are the DNMT genes that have been shown to have a role in the establishment and regulation of cytosine methylation (Bird 2002). Another possible interpretation of the model is that the switching rate is a mutation rate between the two A/a alleles at the major locus under selection. The evolution of mutation has been an area of intense research in population genetics and exceptions to the Feldman–Liberman reduction principle (Feldman and Liberman 1986) are expected in populations experiencing spatial and temporal variation. Because the B/b modifier locus may be genetic or epigenetic, we explore a wide range of switching rates.
There are four possible genotypes in the population AB, Ab, aB, and ab. At each generation, the population goes through recombination, selection, and phenotypic switching, in each deme separately, and then individuals may migrate between the two demes. As in general analyses of neutral modifiers, we frame the question in terms of the stability of the state of fixation in B, producing switching rate μB, to invasion by allele b, which produces switching rate μb. We assume these rates of switching to be the same in both phenotypic directions. This symmetry assumption makes our analysis more tractable but relaxing this assumption should not change the general conclusions of the model (see Salathé et al. 2009). In particular, we ask how the migration between Ex and Ey, which is assumed to occur at rate m per generation, affects this stable switching rate. We shall see that much of the analytical structure of the mathematical problem recapitulates that seen in Balkau and Feldman (1973), where there was no switching from A to a but where locus B/b controlled the migration rate between Ex and Ey with constant fitnesses in the two demes.
We first present the case in which selection is constant in the two demes. We find that the reduction principle (zero is the uninvadable switching rate) holds for all possible migration rates. We then consider the case in which there is periodic fluctuating selection within each of the demes. In this case, we find parameter regimes in which the reduction principle does not hold, and nonzero switching rates can be favored. These evolutionarily stable rates are shown to depend both on the environmental volatility and on the migration rate of the system.
Constant Selection
Let the frequencies and the fitnesses of the four genotypes in the two demes be as follows:
| (1) |
As the modifier locus is selectively neutral, we have
| (2) |
With allele B at the modifier locus, the switching rate in both directions between alleles A and a in each deme is μB, and it is μb with the modifier allele b. We denote by r the recombination rate between the two loci in both demes and by m the rate of migration between the demes.
Thus, in the next generation, after recombination, selection, phenotypic switching, and migration, the new frequencies and of the genotypes in the two demes are
| (3) |
where
| (4) |
with
| (5) |
| (6) |
This general model is very complicated and, to gain some mathematical insight, we assume the following symmetry relations among the fitness parameters:
| (7) |
Thus, we assume that with s > 0, phenotype A is preferred in deme Ex, while phenotype a is preferred in deme Ey and there is symmetry in these fitness differences between the demes (Ishii et al. 1989; Lachmann and Jablonka 1996). Symmetry assumptions in selection pressures are made throughout this article and are important for the mathematical tractability of the model. Future work will focus on the effect of relaxing these symmetry assumptions on the general conclusions of the model.
Boundary equilibria with symmetric selection
We start our analysis by assuming that initially only allele B is present at the modifier locus; i.e., x2 = x4 = y2 = y4 = 0. To simplify the notation, since x3 = 1 − x1 and y1 = 1 − y3, let x1 = x and y3 = y. With this notation, the change in the population state following the transformations given in (3) and (4) becomes
| (8) |
Here,
| (9) |
We first study the equilibrium points of (8). With 0 < m, μB < 1, (8) has no equilibria where one of the two alleles A or a is fixed in either deme (i.e., x = 0, x = 1, y = 0, y = 1 are impossible). To find the polymorphic equilibria, we examine the change in (y − x). A simple computation shows that
| (10) |
Hence, at equilibrium, either y = x or
| (11) |
We call an equilibrium with y = x a “symmetric equilibrium” and any equilibrium with y ≠ x, such that (11) holds, an “asymmetric equilibrium.”
Let us write
| (12) |
The following result holds when 0 < m, μB < 1:
Result 1.
-
On the boundary, where only allele B is present, there is a unique symmetric equilibrium (x∗, y∗) with
(13) - where x∗ is the unique positive root of the quadratic equation
(14)
(x∗, y∗) is internally stable on the boundary with only B present.
If 0 < m, then
The proof of this result is in Supporting Information, File S1.
In addition to the symmetric equilibria, asymmetric equilibria may exist when (11) is satisfied. In fact, we have the following result:
Result 2. If 0 < m < m0 and 0 < μB < μ0, where
| (15) |
then there is a unique asymmetric equilibrium Here,
| (16) |
where is the unique positive root of the quadratic equation
| (17) |
and
| (18) |
A proof of Result 2 can be found in File S2.
An interesting question is whether the asymmetric equilibrium can be internally stable. The mathematical analysis of this issue is complicated, but we conjecture that the asymmetric equilibrium is never internally stable in the boundary with only the B allele present. Our conjecture is based on extensive simulations of the model and the following result, which holds when the switching rate μB is small.
Result 3. When μB is “small”, the asymmetric equilibrium is “near” the fixation of B in the two demes and is internally unstable.
See File S3 for the proof of this result.
External stability of the symmetric equilibrium
We now check the local stability of the symmetric equilibrium (x∗, y∗) to the introduction of the modifier allele b, which changes the switching rate from μB to μb. To this end, we investigate the linear approximation L∗ to the transformation of the population state near (x∗, y∗), such that, up to nonlinear terms,
| (19) |
Here, ε = (ε1, ε2, ε3, ε4), δ = (δ1, δ2, δ3, δ4) with εi and δi small, and and similarly for ε′ and δ′.
The stability of (x∗, y∗) is determined by the eigenvalues of L∗. This approach has been used extensively in modifier theory (see, for example, Feldman and Liberman 1986). The matrix L∗ has the structure
| (20) |
where we have swapped the columns and the rows. The matrix ∗ does not affect the eigenvalues of L∗, which are those of the two submatrices Lin and Lex. Lin determines the internal stability of (x∗, y∗) in the boundary where only B is present. Since (x∗, y∗) is assumed to be internally stable, the eigenvalues of Lin are less than 1 in magnitude. Lex corresponds to the linear approximation of the evolution near (x∗, y∗) of the genotypes Ab and ab in the two demes.
A straightforward computation shows that the 4 × 4 matrix Lex can be written as
| (21) |
where A, B, C, D are positive and depend on (x∗, y∗) and on s, r, and μb. Lex has exactly the same form as the matrix that appears in the external stability analysis of Balkau and Feldman (1973). What enables us to analyze the positive largest eigenvalue of Lex, which determines the external stability of (x∗, y∗), are the following properties of matrices L of the form (21):
-
If P is the 4 × 4 matrix given by
(22) - then
(23) - where 0 is the 2 × 2 zero matrix and
(24) Thus L is “similar”, via P, to the block matrix (23).
-
2.The fourth degree characteristic polynomial of L factors into the product of the two quadratic characteristic polynomials of M and N. These two polynomials share the same constant term
(25) -
3.In general if, for fixed m, L1, L2, … , Ln are matrices of the form (21) with corresponding positive entries Ai, Bi, Ci, Di for i = 1, 2, … , n, then
(26) - Here Mi and Ni are the M and N matrices of (23) associated with Li. Moreover, the fourth-degree characteristic polynomial of factors into the two second-degree characteristic polynomials of and . These two polynomials share the same constant term
(27)
In File S4 we use properties 1 and 2 to obtain the following external stability result.
Result 4. When 0 < m, μB, the symmetric equilibrium (x∗, y∗) is externally stable to the introduction of the new modifier allele b if μb > μB and is unstable if μb < μB. Therefore, the reduction principle holds and zero is the only uninvadable switching rate.
Overview of results when selection pressures are constant
Intuitively, in this model, both migration and phenotypic switching conspire to reintroduce the deleterious phenotype in each of the two spaces, which selection is trying to remove. Therefore, it is expected that both the migration and the switching rates evolve toward zero (Balkau and Feldman 1973; Liberman and Feldman 1986). For a fixed migration rate, as presented here, phenotypic switching will not evolve, and the population will approach a migration–selection balance at the major loci.
Periodically Fluctuating Selection
Suppose that within each deme the selection regime varies temporally. We assume periodically fluctuating selection and explore the evolution of the switching rate as a function of both the migration rate and the length of the period of the fluctuating selection, which we also refer to as the environmental period. Specifically, we assume two types of selection regimes, type 1 with the fitness parameters
| (28) |
and type 2 with
| (29) |
In the general case, an environmental cycle consists of (k + ℓ) phases of selection, the first k of type 1 followed by ℓ phases of type 2 selection. We first mathematically explore the dynamics of the system in the cases k = ℓ = 1 (cycle of period 2) and k = ℓ = 2 (cycle of period 4). These are the cases in which we are able to derive analytical results. We then study the system in the case of larger environmental periods, using numerical approximations and simulation.
Assumptions and limitations of the fluctuating selection model
For analytical tractability, we make stringent symmetry assumptions on the fitnesses of the two phenotypes A and a, both in time and in space. We assume that, within each environment, each deme has an optimal phenotype and there is symmetry in these fitness differences between the demes. We also assume that, within each deme, each temporal environment produces a different optimal phenotype and, again, symmetry in these fitness differences is assumed. Another important assumption of the model is that there are only two different environments that fluctuate periodically. We do not expect the results to change qualitatively if there are more than two environments; we also hypothesize that introducing random waiting times for environmental change will affect the evolutionarily stable switching rate, but not whether nonzero switching rates can evolve (Salathé et al. 2009). Moreover, it is known that asymmetry in selection pressures can substantially affect evolved switching rates in a model with just temporal heterogeneity (Salathé et al. 2009). Therefore, future work will need to focus on the effect of relaxing these symmetry assumptions on the conclusions of our model.
Cycle of period 2 (k = ℓ = 1)
Here, selection goes through two phases. The first phase is type 1 as in (28) and the second is type 2 as in (29).
Again, we first study the boundary equilibria where only the modifier allele B is present and the switching rate is μB. The initial frequencies of AB in the two demes are denoted by x and y, respectively. After phase 1, they change to and and after phase 2 they become x′ and y′, the frequencies at the start of the next cycle. We have
| (30) |
and
| (31) |
Due to the symmetry in the system, we have
| (32) |
Hence,
| (33) |
Thus, at equilibrium where (y′ − x′) = (y − x), we have either y = x or
| (34) |
Equilibria with y = x are the symmetric equilibria, while if y ≠ x and (34) holds, we have asymmetric equilibria. For the symmetric equilibria we have the following:
Result 5. When 0 < m, μB < 1, and s > 0, a unique symmetric equilibrium exists such that
| (35) |
is the unique positive root of R(x) = 0, where
| (36) |
with mB = m + μB − 2mμB. If, in addition, , then .
The proof of this result is presented in File S5. In File S6 we also show the following:
Result 6. The unique symmetric equilibrium is internally stable on the boundary where only the modifier allele B is present.
For the local stability of this symmetric equilibrium to the introduction of the “new” modifier allele b, the external stability matrix Lex is given by
| (37) |
where both and have the form (21). Thus, using the properties of these matrices, the characteristic polynomial of Lex factors into two quadratic polynomials, which allows us to find the conditions needed for external stability. The complete analysis is given in File S7, where the following result is proved:
Result 7. In the case of selection fluctuating with period 2, the internally stable unique symmetric equilibrium is externally stable when μB > μb and is unstable when μB < μb, for all 0 ≤ r ≤ 1 and Thus, in this case, higher switching rates are favored, and the evolutionarily stable phenotypic switching rate is 1.
The mean fitness and external stability
We saw that in a constant environment the symmetric equilibrium is externally stable if μb > μB and unstable if μb < μB, so that smaller switching rates are favored. On the other hand, with fluctuating selection of period 2, nonzero switching rates evolve; the symmetric equilibrium is stable if μB > μb and unstable if μB < μb. This phenomenon is related to the behavior of the mean fitness at equilibrium in the following way:
Result 8. The mean fitness at the symmetric equilibrium is (i) a decreasing function of μB in a constant environment or (ii) an increasing function of μB in a period 2 cycling environment.
The proof is given in File S8.
Cycle of period 4 (k = ℓ = 2)
In this case, in each generation, the fitnesses of the genotypes in each population go through four phases (“1”, “2”, “3”, “4”). The fitnesses at these phases are
| (38) |
The analysis of this case for r = 0 parallels that of Liberman et al. (2011) if we replace μM by mB = m + μB − 2mμB and μm by mb = m + μb − 2mμb (see Result 5). Our results for this case can be summarized as follows.
On the boundary where B is fixed, if we assume that x1 = y3, x3 = y1 through the four phases of a cycle, then the four-step recursion is
| (39) |
where x = (x1, x3) = (y3, y1), and T1 and T2 are the transformations associated with fitness “1” and fitness “3”, respectively. The mean fitness , which is the normalizing factor, is
| (40) |
If r = 0, we can show that, at the symmetric equilibrium we have Therefore, at equilibrium,
For the external stability of note that if mb = 0, the matrices governing the evolutionary dynamics of the system in steps T1 and T2 become
| (41) |
respectively. If mb = 1, the matrices in steps T1 and T2 become
| (42) |
respectively. As a result, when mb = 0, the local stability is governed by the matrix M∗:
| (43) |
and when mb = 1,
| (44) |
Since in the case r = 0 is externally stable when either mb = 0 or mb = 1. Since mb = 0 if m = 0, μb = 0, or m = 1, μb = 1 and mb = 1 if m = 0, μb = 1 or m = 1, μb = 0 (which is not biologyically relevant), we conclude that when both r and m are small for any 0 < μb < 1, a new mutation-modifying allele cannot invade if it produces switching rates that are too high or too low. This suggests that there might be an intermediate switching rate μB that gives rise to a symmetric equilibrium which cannot be invaded by allele b. Actually, we have the following result:
Result 9. In the period 4 symmetric case, if r = 0, the mean fitness achieves a maximum at
The proof is similar to that in Liberman et al. (2011). Note that since mB = m +μB(1 − 2m), implies that (1 − 2m)(1 − 2μB) = 0, and as coincides with Hence, for fixed has its maximum when The next result is based on extensive simulations.
Result 10. In the period 4 symmetric case with 0 ≤ r ≤ 1 and for all migration rates, the phenotypic switching rate cannot be invaded by rates either smaller or larger than Thus, is the stable switching rate.
Here, again, the migration rate does not affect the uninvadable switching rate. Moreover, with a switching rate of 1/2, both alleles A and a reach a frequency of 0.5 in each deme, and thus migration acts to neutrally reshuffle these alleles only between the two demes.
Cycle of period k + ℓ
Following our previous analyses where k = ℓ = 1 and k = ℓ = 2, we conjecture that, in the general case, on the boundary, where the modifier allele B is fixed, we have the following result:
Result 11. If 0 < m, μB < 1, and s > 0, a unique symmetric equilibrium exists such that
| (45) |
and is the unique positive root of a quadratic equation
| (46) |
with G(0) < 0 and G(1) > 0, where p and q are functions of s, μB, k, and l. Moreover, this symmetric equilibrium is internally stable.
Our conjecture is based on analytical examination of several choices of the (k + ℓ) phases, using Mathematica, and on extensive simulations.
The external stability analysis of is very complicated in the general case with arbitrary k and ℓ, as the associated external stability matrix Lex is then
| (47) |
corresponds to selection phase i in the k phases where selection is of type 1, and is associated with phase j of the ℓ phases where selection is of type 2.
Each of these (k + ℓ) matrices is of the form
| (48) |
with corresponding parameters for and for (similar to the matrices presented in File S7). All the matrices and for i = 1, 2, … , k and j = 1, 2, … , ℓ are positive.
Applying the properties of matrices of the form (48), we have the following result:
Result 12. The characteristic polynomial of Lex factors into two quadratic polynomials. The constant terms of both polynomials are
| (49) |
The constant term (49) is always positive, and based on extensive mathematical checks using Mathematica it is always <1. Therefore, using the same argument as in the case k = ℓ = 1, the external stability of the symmetric equilibrium is determined by a sole condition C1(1) > 0 where C1(λ) is the “first” quadratic polynomial in the factorization of the characteristic polynomial of Lex.
Overview of analytic results when selection pressures are periodic, periods 2 and 4
When environments change periodically, the reduction principle no longer holds and nonzero switching rates are evolutionarily stable and uninvadable. In fact, if the environment changes every generation or every two generations, this evolutionarily stable switching rate does not depend on the migration rate in the system and is 1/n as reported by previous studies (where n, the number of generations in each environmental phase, is 1 or 2, respectively). An important result is that this evolutionarily stable switching rate maximizes mean fitness at equilibrium. This result is deeply rooted in classic population genetics theory and is connected to early work on neutral modifiers and the mean fitness principle of Karlin and McGregor (1972).
We were not able to derive mathematical expressions for the external stability conditions for cycles k = l > 2. For these longer cycles of equal period (which we from now on denote by 2n, where n is the number of generations in each phase), we use simulation and numerical analyses and find that the leading eigenvalue depends not only on the difference between μB and μb, but also on the initial switching rate μB and the migration rate. We present these results in the next section.
Simulations and Numerical Analyses
Description of the simulation
We start with a population fixed on the B allele at the modifier locus, which determines a switching rate μB sampled randomly between 0 and 1 and then held constant. The population evolves for 1000 generations until it reaches an internally stable symmetric equilibrium at the A/a locus. We then introduce the b allele at a small frequency (10−4) near this symmetric equilibrium. The switching rate μb set by allele b is chosen as the product of the resident switching rate and a number generated from an exponential distribution with mean 1. This allows us to test invading mutation rates that are more often close to the resident rate, but also rates that are far from it. After 5000 generations with these fixed values of μB and μb, we determine whether the newly introduced allele was able to invade in the population; if the frequency of b is larger than its initial frequency of 10−4, we declare invasion. This is the initial invasion trial. If the b allele was able to invade, we start the next invasion trial with this invading switching rate chosen as the new resident switching rate of the B allele. If there was no invasion, the B allele determines the same resident switching rate as the previous trial. We then repeat the invasion steps described above. After at least 500 invasion trials and after the resident switching rate cannot be invaded in 50 consecutive trials, the final switching rate in the population is declared to be the stable switching rate.
Results
The case in which each environment persists for three generations before a change shows some anomalies is presented in Figure S1 as a function of the migration and recombination rates. In the case of no migration, Ishii et al. (1989) first showed that there are two equilibria for the mutation rate, one stable and one unstable. It was also shown that increasing the recombination rate r can change the stability and the domains of attraction of the two equilibria. With positive migration, we show that the uninvadable switching rate is sensitive to the interplay of recombination and migration, with possible sharp discontinuities in the stable switching rate. For no recombination and small migration rate, any switching rate can be invaded by a larger one. As the recombination rate increases, for small migration, the stable mutation rate is on the order of 1/3. As the migration rate increases, the stable switching rate decreases in a manner consistent with larger values of n.
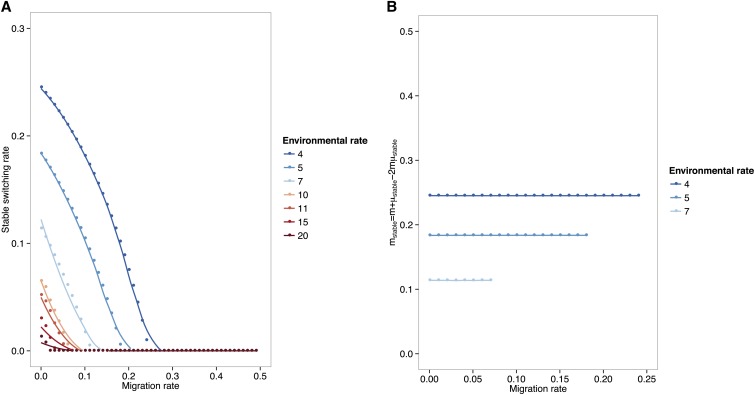
The results for the evolutionarily stable switching rate as a function of migration rate for periods 2n > 6 are presented in Figure 1A. The stable switching rate is seen to be a decreasing function of both the migration rate and the number of generations n before an environmental change. Indeed, we confirm that, in the absence of migration, the stable switching rate is of the order of 1/n as reported by previous studies. As the migration rate increases, the stable switching rate decreases until, above a threshold migration rate, it becomes zero. Note that this threshold in migration also is on the order of 1/n. This is intuitive, since in this case where the environments both change at the same time, a migration rate inversely proportional to the rate of environmental change would place the migrants in a deme and environment with the same selection pressure as the one to which they were adapted. This effectively eliminates the need for switching. For migration rates below this threshold, the uninvadable switching rate is such that, together with migration, it generates the required variability that allows the system to cope with the environmental change it experiences. In fact, Figure 1B shows that the stable switching rate is such that the parameter mstable = m + μstable − 2mμstable (the equivalent of mb in Result 5) is of the order 1/n.
Figure 1.
The evolutionarily stable switching rate as function of migration and number of generations before an environmental change, n > 3. (A) The symmetric selection coefficient s = 0.4. Recombination rate is r = 0.2. We vary the rate of environmental change n as in the legend. The dots represent the average over 10 runs of the simulation. The plotted curves represent a fit to the data using a generalized additive model with penalized cubic regression splines. The stable switching rate is a decreasing function of both the migration rate and the rate of environmental change. (B) The parameter mstable = m + μstable − 2mμstable. The symmetric selection coefficient s = 0.4. Recombination rate is r = 0.2. The rate of environmental change n varies as in the key. The dots represent the average over 10 runs of the simulation.
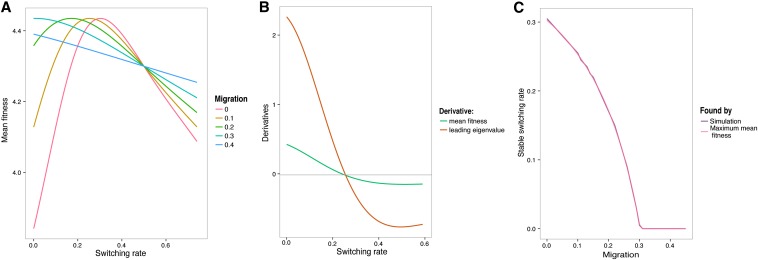
In Figure 2 we show that, for recombination rate r = 0, there is an alternative way to obtain the evolutionarily stable switching rate. Ishii et al. (1989) found that, when r = 0, the evolutionarily stable switching rate is the one “that maximizes the long-term geometric average of population fitness.” Therefore, we expect that the switching rate that maximizes mean fitness at equilibrium is also the uninvadable switching rate (also see Result 8). The invasibility condition is again given by the properties of the leading eigenvalue λ0 of the local stability matrix of the system. To determine invasibility numerically, we can differentiate λ0 with respect to μb and then evaluate this derivative at μb = μB, where we know that λ0 = 1. The sign of this derivative then tells us whether a larger or a smaller value of μb will produce a leading eigenvalue λ0 larger or smaller than 1 and thus whether b will invade or not. Therefore, the values of μb where this derivative crosses zero with a negative slope correspond to values of μB that are uninvadable. We see that the switching rates that maximize the mean fitness at equilibrium (and are zeros for the derivative of mean fitness with respect to μB) are also the zeros of the derivative of λ0 and therefore can be regarded as uninvadable. Figure 2B shows this equality of the critical points for the two derivatives, using an example where the environment changes every n = 4 generations, the symmetric selection coefficient is s = 0.4, and the migration rate m = 0.1. Figure 2C demonstrates the match between the stable, uninvadable switching rates found by simulation and the switching rate that maximizes mean fitness at equilibrium.
Figure 2.
The link between the evolutionarily stable switching rate and the switching rate that maximizes mean fitness at equilibrium. The symmetric selection coefficient s = 0.4. Recombination rate is r = 0. The environment changes every n = 4 generations. (A) Mean fitness at equilibrium, for the range of migration rates in the key. In B, using the migration rate m = 0.1 as an example, we show that the switching rate that maximizes mean fitness at equilibrium is also the switching rate that is uninvadable by other rates; in other words, the zeros of the derivative of the mean fitness at equilibrium are also the zeros of the derivative of the leading eigenvalue of the local stability matrix. (C) The match between the stable switching rates found by simulation and the switching rates that maximize mean fitness at equilibrium, as a function of the migration rate.
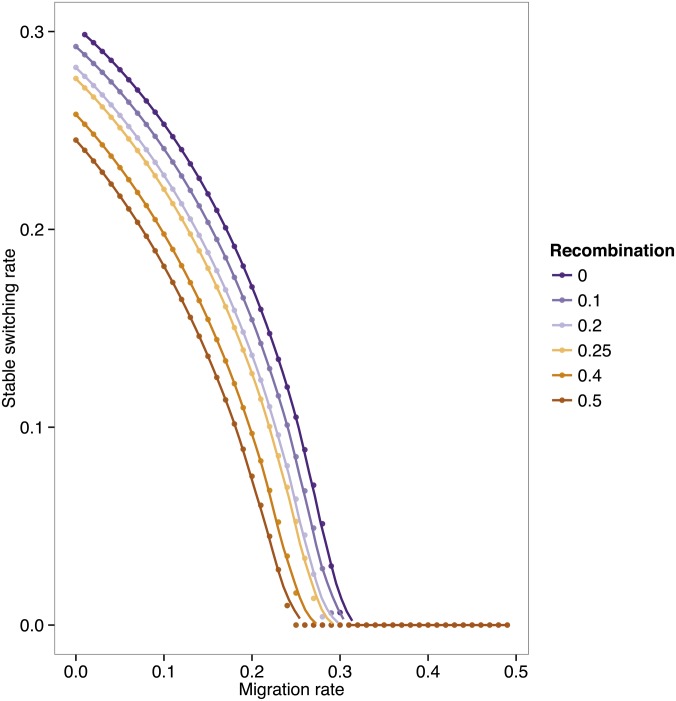
Figure 3 shows the stable switching rate when the environment alternates every four generations as a function of the migration and recombination parameters. We find that the stable switching rate is a decreasing function of the recombination rate, for any migration rate (see also Liberman et al. 2011). As recombination increases, the force of secondary selection on the modifier decreases as the linkage disequilibrium between the two loci decreases. This is expected to be a relatively weak force, which explains the relatively small difference between the curves in Figure 3 compared to the effect of migration (see Figure 1) for a given recombination rate. In Figure S2, the effect of increasing period is also seen to be more pronounced than that of increasing the recombination rate. In earlier studies of neutral modifiers of mutation and recombination, the induced selection on the modifier locus was of the order of the square of the disequilibrium between the major and the modifier loci. Even if this effect is exaggerated somewhat by the environmental fluctuations, it remains weaker than the effect of either the period of fluctuation or the migration rate.
Figure 3.
The evolutionarily stable switching rate as a function of migration and recombination. The symmetric selection coefficient s = 0.4. The environment changes every n = 4 generations. The plotted curves represent a fit to the data using a generalized additive model with penalized cubic regression splines.
In Figure S3, we show that the strength of the symmetric selection coefficient s does not qualitatively influence the evolutionary dynamics of the system. This result is in accordance with previous analyses in temporally varying environments showing that changing the symmetric selection coefficient does not affect the evolutionarily stable switching rate (Ishii et al. 1989; Salathé et al. 2009).
Discussion
In recent years, the evolution of microbial populations has become a question of significant biological importance. The medical threat posed by the ability of some bacterial species to resist antibiotics by using phenotypic heterogeneity (Dubnau and Losick 2006; Smits et al. 2006) is significant (Centers for Disease Control 2013). Stochasticity and growth bistability allow bacteria to create diverse phenotypes and specialized cells in anticipation of possible adverse changes in the selection pressures (Barrett Deris et al. 2013). For example, bacterial persistence is a well-known mechanism that depends on stochastic switching between normal and persister phenotypes, which are able to survive better in the presence of antibiotics but are virtually growth arrested in standard media (Balaban et al. 2004). Given the role that stochastic switching has in bacterial populations, there is a need to consider the kinds of selective pressures that promote the evolution of switching behaviors and may be associated with the efficacy and evolvability of drug resistance.
We have investigated the evolution of stochastic switching under both temporal and spatial volatility, using a population genetic model with a modifier gene that controls the rate of switching (for examples see Smits et al. 2006; Veening et al. 2008; Beaumont et al. 2009; Rainey et al. 2011). We study conditions under which switching is adaptive and determine the evolutionarily stable (i.e., uninvadable) switching rates for a wide range of migration rates, recombination rates, symmetric selection, and periodic rates of environmental fluctuation. Closed-form analytical results such as those presented here help in gaining insight into the driving evolutionary process that we can then use to inform experimental and possibly medically motivated studies.
Our model reproduces the results found in previous studies with no spatial heterogeneity, but the migration rate between demes with different selection pressures can dramatically alter the evolutionarily stable switching rate. Our results demonstrate that the stochastic phenotypic switching rate is a decreasing function of the migration rate between demes. Moreover, with both spatial and temporal heterogeneity in the system, for environmental periods 2n > 6, there exists a threshold migration rate above which stochastic switching cannot evolve. This threshold is also of the order of 1/n, where n is the number of generations spent in each environment. Below this threshold, the switching rate evolves to the order of 1/n minus the migration rate in the system. We also find that the switching rate is a decreasing function of the recombination rate in the system, although this effect is small compared to that of the migration rate.
Our work highlights the importance of incorporating spatial heterogeneity when studying stochastic switching in the laboratory. Thus space-dependent fitness conditions ought to play a central role in the design and interpretation of experiments that seek to understand the behavior of natural populations and should try to recreate the natural time and space variations to which these populations would be subject. Populations of cells or strains of bacteria in the wild can experience both temporally and spatially heterogeneous selection pressures, and understanding the different contributions of these two inherently different selection regimes may help to interpret the differences observed between wild and laboratory populations (Dubnau and Losick 2006). Moreover, current methods of studying drug efficacies are often performed in bulk growth conditions, which might fluctuate spatially in unforeseen ways. A better understanding of the role of phenotypic variability in these populations could enable the development of new techniques and treatment strategies against drug-resistant bacteria.
Although this work is framed in terms of a gene controlling phenotypic switching, the model is general enough to be applicable to a wider range of dynamical systems. The evolutionary dynamics of the modifier locus B/b can provide insight into the evolution of an epigenetic locus that is able to influence the transition between two different phenotypes, a locus controlling the mutation rate between the two alleles at the major locus A/a, or the rate of switching between two expression levels.
Here we have focused exclusively on the case in which the environment in the two demes varies periodically, with equal numbers of generations before environmental changes in each deme. We have also studied only the case of symmetric selection, where the selection coefficients in the two demes are equal. Previous work in temporally fluctuating environments (Salathé et al. 2009; Liberman et al. 2011) has shown that asymmetry in selection pressures or variance in times between environmental changes can substantially change the optimal switching rate. Incorporating selection asymmetries and variability in the environmental fluctuations is an important follow-up to this work.
The question of optimal phenotypic variability and the evolution of stochastic switching needs to be further explored under more general ecological scenarios driven by both temporal and spatial change. Using both analytic and numerical arguments we have made a first step in this direction: we find that under spatial and temporal fluctuations in selection pressures, phenotypic switching can evolve only when migration is not too high, and its evolution is heavily controlled by the forces driving variability in the population: migration, recombination, and the rate of change in selection pressure.
Supplementary Material
Acknowledgments
Research supported in part by the Center for Computational Evolution and Human Genomics at Stanford. The authors thank Rob Furrow and two anonymous reviewers for helpful comments on the manuscript.
Footnotes
Communicating editor: L. M. Wahl
Literature Cited
- Acar M., Becskei A., van Oudenaarden A., 2005. Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228– 232. [DOI] [PubMed] [Google Scholar]
- Acar M., Mettetal J. T., van Oudenaarden A., 2008. Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet. 40: 471– 475. [DOI] [PubMed] [Google Scholar]
- Arnoldini M., Mostowy R., Bonhoeffer S., Ackermann M., 2012. Evolution of stress response in the face of unreliable environmental signals. PLoS Comput. Biol. 8(8): e1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban N. Q., Merrin J., Chait R., Kowalik L., Leibler S., 2004. Bacterial persistence as a phenotypic switch. Science 305: 1622–1625. [DOI] [PubMed] [Google Scholar]
- Balkau B. J., Feldman M. W., 1973. Selection for migration modification. Genetics 74: 171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont H. J. E., Gallie J., Kost C., Ferguson G. C., Rainey P. B., 2009. Experimental evolution of bet hedging. Nature 462: 90–94. [DOI] [PubMed] [Google Scholar]
- Bird A., 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16: 6–21. [DOI] [PubMed] [Google Scholar]
- Bonduriansky R., Day T., 2009. Nongenetic inheritance and its evolutionary implications. Annu. Rev. Ecol. Evol. Syst. 40: 103–125. [Google Scholar]
- Carja O., Liberman U., Feldman M. W., 2013. Evolution with stochastic fitnesses: a role for recombination. Theor. Popul. Biol. 86: 29–42. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control, 2013 Antibiotic resistance threats in the United States. Available at: http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013–508.pdf.
- Danchin E., Wagner R. H., 2010. Inclusive heritability: combining genetic and nongenetic information to study animal culture. Oikos 119: 210–218. [Google Scholar]
- Deris J. B., Kim M., Zhang Z., Okano H., Hermsen R., Groisman A., Hwa T., 2013. The innate growth bistability and fitness landscapes of antibiotic-resistant bacteria. Science 342: 1237435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Losick R., 2006. Bistability in bacteria. Mol. Microbiol. 61(3): 564572. [DOI] [PubMed] [Google Scholar]
- Feldman M. W., Liberman U., 1986. An evolutionary reduction principle for genetic modifiers. Proc. Natl. Acad. Sci. USA 83: 4824–4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrow R. E., Christiansen F. B., Feldman M. W., 2011. Environment-sensitive epigenetics and the heritability of complex diseases. Genetics 189: 1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal B., Pitchford J. W., Wood A. J., 2010. Exact results for the evolution of stochastic switching in variable asymmetric environments. Genetics 184: 1113–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Matsuda H., Iwasa Y., Sasaki A., 1989. Evolutionarily stable mutation rate in a periodically changing environment. Genetics 121: 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., McGregor J. L., 1972. The evolutionary development of modifier genes. Proc. Natl. Acad. Sci. USA 69: 3611– 3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B. B., Yang Q., Mettetal J. T., van Oudenaarden A., 2007. Heritable stochastic switching revealed by single-cell genealogy. PLoS Biol. 5: e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E., Leibler S., 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309: 2075–2078. [DOI] [PubMed] [Google Scholar]
- Lachmann M., Jablonka E., 1996. The inheritance of phenotypes: an adaptation to fluctuating environments. J. Theor. Biol. 181: 1–9. [DOI] [PubMed] [Google Scholar]
- Leibler S., Kussell E., 2010. Individual histories and selection in heterogeneous populations. Proc. Natl. Acad. Sci. USA 107(29): 13183–13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh E. G., 1970. Natural selection and mutability. Am. Nat. 104(937): 301–305. [Google Scholar]
- Liberman U., Feldman M. W., 1986. Modifiers of mutation rate: a general reduction principle. Theor. Popul. Biol. 30(1): 125–142. [DOI] [PubMed] [Google Scholar]
- Liberman U., Feldman M. W., 1989. The reduction principle for genetic modifiers of the migration rate, pp. 111–137 in Mathematical Evolutionary Theory, edited by Feldman M. W. Princeton University Press, Princeton, NJ. [Google Scholar]
- Liberman U., van Cleve J., Feldman M. W., 2011. On the evolution of mutation in changing environments: recombination and phenotypic switching. Genetics 187: 837–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. N., van Oudenaarden A., 2007. A multistep epigenetic switch enables the stable inheritance of DNA methylation states. Nat. Genet. 39(2): 269–275. [DOI] [PubMed] [Google Scholar]
- Mettetal J. T., Muzzey D., Pedraza J. M., E. M. Ozbudak, and A. van Oudenaarden, 2006. Predicting stochastic gene expression dynamics in single cells. Proc. Natl. Acad. Sci. USA 103: 7304– 7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. A., 1992. The evolutionary maintenance of alternative phenotypes. Am. Nat. 139(5): 971–989. [Google Scholar]
- Rainey P. B., Beaumont H. J. E., Ferguson G. C., Gallie J., Kost C., et al. , 2011. The evolutionary emergence of stochastic phenotype switching in bacteria. Microb. Cell Fact. 10: S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salathe M., Van Cleve J., Feldman M. W., 2009. Evolution of stochastic switching rates in asymmetric fitness landscapes. Genetics 182: 1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits W. K., Kuipers O. P., Veening J. -W., 2006. Phenotypic variation in bacteria: the role of feedback regulation. Nat. Rev. Microbiol. 4(4): 259–271. [DOI] [PubMed] [Google Scholar]
- Starrfelt J., Kokko H., 2012. Bet-hedging—a triple trade-off between means, variances and correlations. Biol. Rev. Camb. Philos. Soc. 87: 742– 755. [DOI] [PubMed] [Google Scholar]
- Thattai M., van Oudenaarden A., 2004. Stochastic gene expression in fluctuating environments. Genetics 167: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening J.-W., Smits W. K., Kuipers O. P., 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62: 193– 210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.