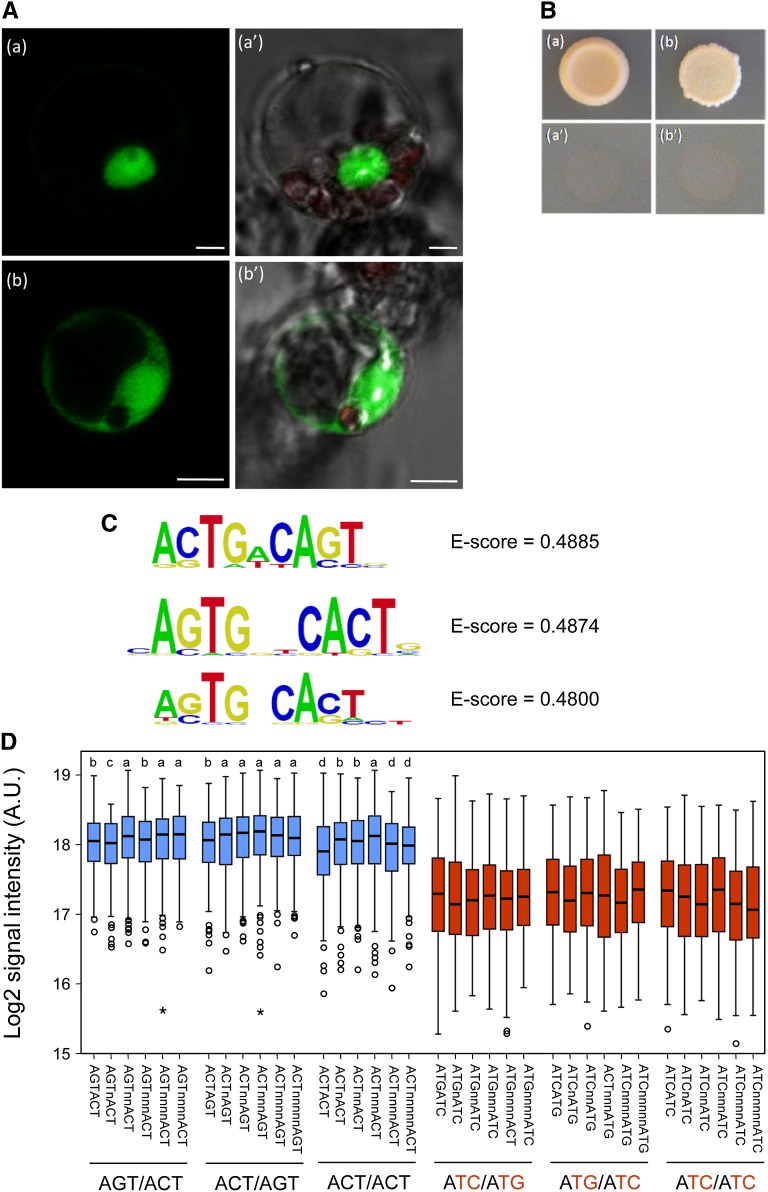
Figure 3.
SlZF2 transcriptional properties. A, Transient expression of YFP-SlZF2 fusion protein in tomato leaf protoplasts: YFP-SlZF2 (Aa and Aa′), YFP control fluorescence (Ab and Ab′), and bright field/chlorophyll/YFP fluorescence respectively. B, SlZF2 transactivation ability. The SlZF2 coding region was fused to GAL4 DNA binding domain (DBD) into the pGBKT7 vector carrying the nutritional marker TRYPTOPHAN1 as the reporter gene. Yeasts were transformed with pGBKT7 empty vector (Ba and Ba′) or with the GAL4-DBD-SlZF2 construct (Bb and Bb′). Both constructs were transformed into the yeast strain Y8930 harboring the ß-galactosidase L, ADE2, and HIS3 reporter genes. Yeasts were separately grown on synthetic dropout medium lacking either Trp (Ba and Bb), or A and His (Ba′ and Bb′). C, Position weight matrix representation of the three top-scoring 8 mers obtained in a seed-and-wobble algorithm. D, Box plot representation of signal intensities of the probes containing the elements indicated. Different combinations of the AGT modules in bipartite motifs are shown in blue, whereas their corresponding mutant versions are shown in red. Letters above the boxes represent different groups of statistical significance, relative to the motif ACTnnnAGT with highest median intensity, as follows: a, P > 0.05 (no significant differences); b, P < 0.01; c, P < 0.001; and d, P < 0.0001 (Wilcoxon exact test). All of the mutant derivatives grouped together at the same significance group (P < 2.2e-16). The number of probes containing the elements indicated ranges from 243 to 303.