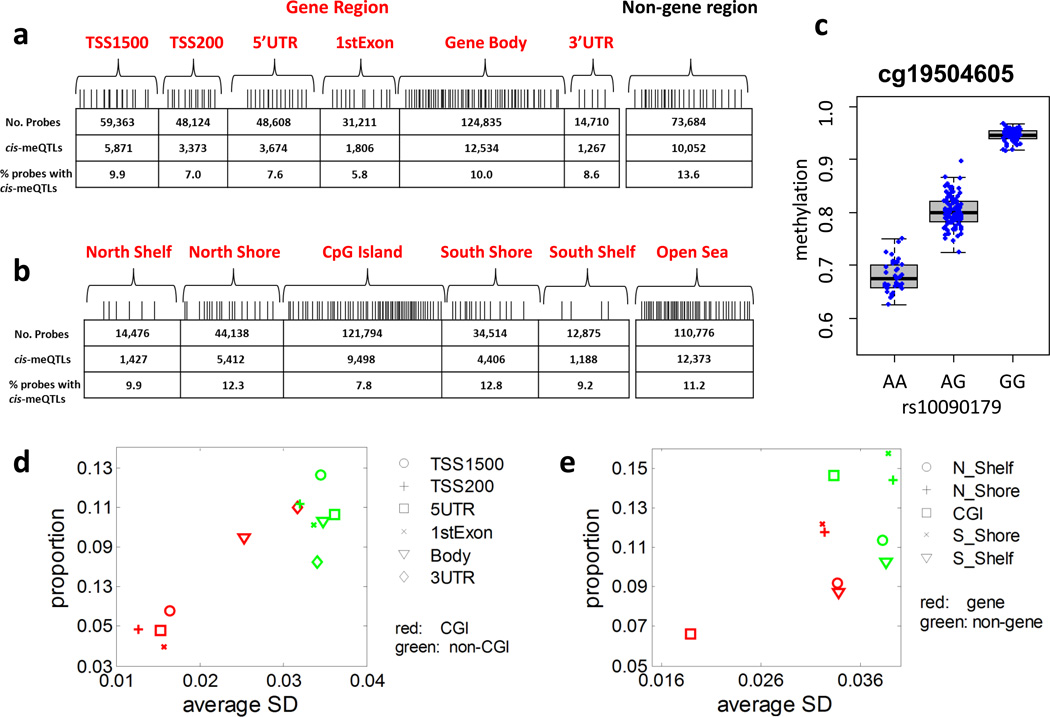
Figure 1. cis-meQTL structural characteristics.
(a) Distribution of CpG probes and corresponding cis-meQTL numbers and proportions in gene and non-gene regions. meQTLs were detected based on EAGLE lung normal tissue samples (n=210). (b) Distribution of CpG probes and corresponding cis-meQTL numbers and proportions in CpG islands (CGIs), shores (< 2kb from the boundary of CGI), shelves (2–4kb from the boundary of CGI) and the remaining region or “open sea”. The box plots show the distribution of the methylation levels in each genotype category with error bars representing the 25% and 75% quantiles. (c) The strongest cis-association is between SNP rs10090179 and CpG probe cg19504605. P=1.5×10−73, t-test. The SNP explains 79.8% of the phenotypic variance. (d, e): The x-coordinate is the average standard deviation (SD) of methylation levels for CpG probes in each category. The y-coordinate is the proportion of CpG probes detected with cis-meQTLs. The proportion of methylation probes detected with cis-meQTLs varied across categories, ranging from 4.0% for CGIs in 1st Exons to 15.7% for south shores in non-gene regions.