Abstract
A capillary with a pulled tip, densely packed with silica particles of 0.47 μm in diameter, is shown to provide higher peak capacity and sensitivity in the separation of intact proteins by reversed-phase liquid chromatography–mass spectrometry (LC–MS). For a C18 bonded phase, slip flow gave a 10-fold flow enhancement to allow for stable nanospray with a 4-cm column length. Model proteins were studied: ribonuclease A, trypsin inhibitor, and carbonic anhydrase, where the latter had impurities of superoxide dismutase and ubiquitin. The proteins were well separated at room temperature with negligible peak tailing. The peak capacity for ubiquitin was 195 for a 10-min gradient and 315 for a 40-min gradient based on Gaussian fitting of the entire peak, rather than extrapolating the full-width at half-maximum. Separation of a cell lysate with a 60 min gradient showed extremely high peak capacities of 750 and above for a peptide and relatively homogeneous proteins. Clean, low noise mass spectra for each model protein were obtained. The physical widths of the peaks were an order of magnitude narrower than those of conventional columns, giving increased sensitivity. All proteins except ubiquitin exhibited significant heterogeneity apparently due to multiple proteoforms, as indicated by both peak shapes and mass spectra. The chromatograms exhibited excellent reproducibility in retention time, with relative standard deviations of 0.09 to 0.34%. The results indicate that submicrometer particles are promising for improving the separation dimension of LC in top-down proteomics.
Proteomics has been developing rapidly over the past 15 years since its inception,1 being extensively used in biomarker discovery and drug development.2,3 In top-down proteomics, mass analyzers with ultrahigh resolution are now available, such as ion cyclotron resonance, Orbitrap, and time-of-flight, with which the molecular weight of intact proteins can be identified with sufficient accuracy to distinguish multiple proteoforms, which arise from post-translational modifications and other processes.4 The ability to fragment proteins in the mass spectrometer enables identification of the positions of post-translational modifications,5,6 which provides information about disease processes.7−10 For top-down proteomics, efficient separation of the protein sample prior to mass spectrometry is critical because biological samples contain proteins with concentrations ranging over many orders of magnitude.11 The efficiency of protein separations presents a major analytical challenge.12−15
Reversed-phase liquid chromatography is typically used as at least one dimension in top-down proteomics because it is automated and easily interfaces with mass spectrometry (MS). The advances in column performance have been relatively slow: for small-molecule separations, the minimum plate heights of the three major types of reversed-phase columns, sub-2 μm, monolithic, and porous shell, are all within a factor of 2 of one another, with the sub-2 μm columns giving a moderately better efficiency.16 For protein separations, it is difficult to compare peak capacities across laboratories because researchers choose different proteins, which have different levels of heterogeneity. A commercial protein is generally quite impure, consisting of multiple proteoforms. Fekete et al. used the same protein, myoglobin, to compare many columns, showing that the best sub-2 μm column performs comparably to the best core–shell column.17 Specifically, using ultraviolet detection, peak capacities as high as 200 and 370 were measured for gradients of 10 and 40 min, respectively. Peak capacities are about 2-fold lower with MS detection for a given gradient time13,18 because trifluoroacetic acid must be used much more sparingly with MS detection due to its suppression of ionization.19 Given the complexity of protein samples, columns with higher peak capacities are needed, especially for the faster gradients that are useful for multidimensional separations.
We previously reported that capillaries packed with 0.47 μm particles exhibited unprecedented efficiency in reversed-phase separation of intact proteins when injected by diffusion and detected on-column by fluorescence microscopy.20 The silica particles inside capillary were so homogeneously packed that eddy diffusion was negligible,21 in contrast to capillaries packed with particles on the micrometer scale.22 Slip flow was shown to occur in these capillaries, which gives both an enhanced flow rate and a decreased velocity distribution in the mobile phase.20 Injection using a commercial nano-liquid chromatography (LC) system, combined with gradient elution, showed that the submicrometer particles capillary had a 5-fold higher speed and 2-fold higher resolution than did the commercial column for intact protein separation.23 The performance of these materials has not yet been studied for LC–MS. In this work, we investigate whether pulling the tip of a capillary that is well packed with 0.47 μm particles will allow a sufficient flow rate to give stable nanospray and whether high efficiency is obtained.
Experimental Section
Preparation of Capillaries Packed with Silica Colloidal Crystals with Pulled Tips
The method for preparing silica colloidal crystals was similar to the procedure described before.24 Briefly, 470 nm diameter silica particles (Nanogiant, Temple, AZ) were calcined at 600 °C for 6 h for three times and then annealed at 1050 °C for 3 h. After rehydroxylation in 50/50% (v/v) nitric acid/water overnight, the particles were rinsed with water and ethanol and finally suspended in ultrapure water to get slurry of 30% (w/w). Fused silica capillaries of 100 μm i.d. coated with polyimide (Polymicro Technologies, Phoenix AZ) were conditioned by pumping 0.1 M NaOH for 15 min, followed by pumping water and ethanol separately, each for 10 min. After drying in the vacuum oven for 30 min, the capillaries were filled with slurry prepared previously. The colloidal crystals were formed inside the capillaries by pumping ethanol through the capillaries under sonication. The packed capillaries were kept at room temperature for completely dryness, which usually takes about 3 days. After the capillaries were dry, 1–2 cm long windows were burned on the capillary for pulling tips. Before modification, the capillaries were placed in a 50% humidity chamber for 30 min. The 2% methyl trichlorosilane and 16% n-octadecyltrichlorosilane (Gelest, Morrisville, PA) were dissolved in dry toluene simultaneously, and the packed capillaries were put into this solution at room temperature and allowed to react overnight. After modification, the capillaries were rinsed by pumping toluene through and then put in the oven at 120 °C for 2 h. The nanospray tips were pulled with the P-2000 micropipet puller (Shutter Instrument, Novato, CA) and ready for use. The bonded phase is expected to be burned away, but since the tip is only 500 μm long, no remodification of the silica was performed.
Reagents and Materials
Standard proteins, ribonuclease A from bovine pancreas, trypsin inhibitor from Glycine max (soybean) and carbonic anhydrase from bovine erythrocytes were purchased from Sigma Chemical (St. Louis, MO). Escherichia coli protein sample (lyophilized control sample for use in IEF applications) was purchased from Bio-Rad (Hercules, CA) and dissolved in water. An Amicon spin filter (cutoff 100k) from Millipore (Billerica, MA) was used to remove large species in the sample. HPLC gradient acetonitrile and water were purchased from Sigma (St. Louis, MO) and used all the time.
Nano-LC–MS
Thermo UltiMate 3000 nano LC System was used to provide gradient elution and sample auto injection. A Thermo LTQ Velos mass spectrometer was used as the detector. The flow rate was 200 nL/min. A nano ESI ion source was used, and the spray voltage was 4 kV. For gradient elution, solvent A, water with 0.5% formic acid and 0.02% trifluoroacetic acid (TFA), and solvent B, acetonitrile with 0.5% formic acid and 0.02% TFA, were used. The concentrations of model proteins in the mixture were prepared as follows: ribonuclease A, 0.05 mg/mL; trypsin inhibitor, 0.05 mg/mL; and carbonic anhydrase, 0.01 mg/mL. The injection volume was set to 2 nL, although it is not quantitatively reliable below 20 nL on this instrument. The scan rate of the mass spectrometer was set to 33.3 kDa/s and the resolution (fwhm) was ≤0.6 Da.
Results and Discussion
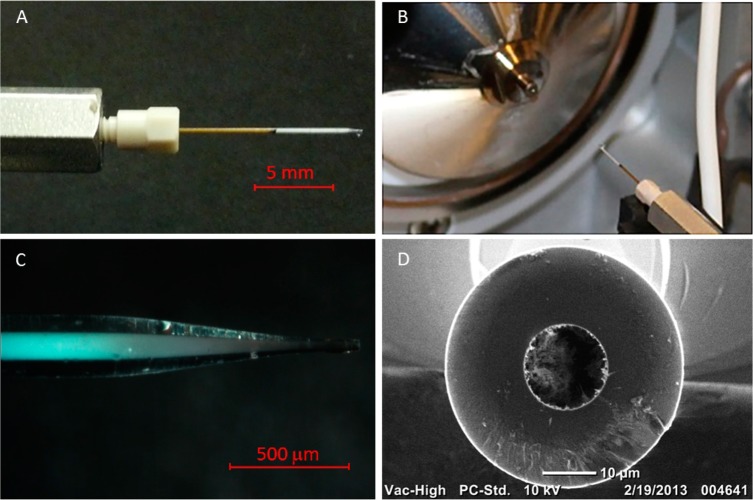
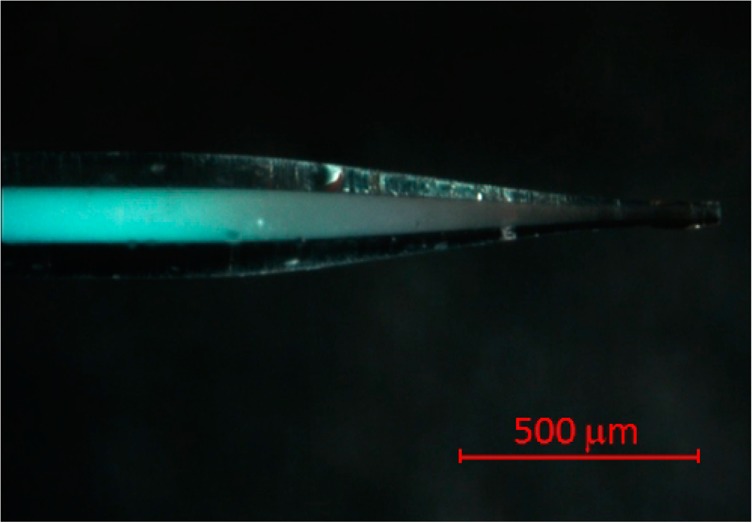
The technology for using these unusually small particle sizes in LC–MS is shown by photographs in Figure 1. A typical capillary connected to the nano-LC system through a commercial union is pictured in Figure 1A, and the interface between the capillary and MS is pictured in Figure 1B, where the metal union provides electrical connection. A column length of 4.0 cm was used, which is slightly shorter than the lengths of commercial RPLC columns, which range from 5 cm and up. This takes advantage of the relative insensitivity on column length for resolution of proteins in gradient elution reversed-phase liquid chromatography, allowing higher flow rates with these submicrometer particles. Coupling capillaries to MS usually employs an emitter with smaller i.d for a lower limit of detection, and the optical image of the pulled capillary tip is shown in Figure 1C. The bluish color is caused by Bragg diffraction, which demonstrates that the particles have face-centered cubic order.24 The packing extends almost to the end of the tip, which minimizes post column broadening.14,15,18 A scanning electron microscope (SEM) image in Figure 1D shows that the tip diameter is 12 μm, which was found to be reproducible within a few micrometers. The SEM image also shows that the particles were not noticeably melted in the tip-pulling process. These pulled capillaries with fritless tips were found to withstand the highest pressure of the instrument, 1 000 bar, avoiding the problem of particle leakage.25 This high stability owes to the particles and walls being polymerized together with trifunctional silanes to form a monolith.
Figure 1.
Column dimensions and MS interface: (A) connection between nano-LC and column, (B) interface between column and MS, (C) microscopy image of column tip, and (D) SEM image of column tip.
The flow properties reveal a 10-fold enhancement from slip flow. Specifically, for the flow rate of 200 nL/min used for the separations in this work, the observed back-pressure for pure water as the mobile phase was no more than 600 bar. On the basis of the Kozeny-Carman equation, which relates back-pressure and flow rate for the case of no-slip,20 the back-pressure is calculated to exceed 7 000 bar. Slip flow enhancement is a nanoscale phenomenon that owes to the attractive interactions between mobile and stationary phases being weak, resulting in a nonzero velocity for the fluid at the wall and consequently an enhancement in flow rate. We previously showed that the 470 nm particle diameter has sufficiently small interstices to give a 5-fold slip flow enhancement for a C4 bonded phase.26 The greater hydrophobicity of the C18 bonded phase in the present work is likely the reason for the 10-fold slip flow enhancement. The slip flow enhancement thus enables stable nanospray for modest UHPLC pressures.
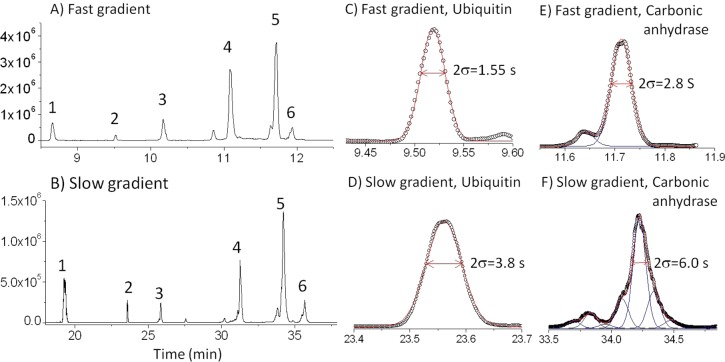
The base peak chromatograms for the three model proteins, ribonuclease A, trypsin inhibitor, and carbonic anhydrase, are presented in parts A and B of Figure 2 for a 10 min (fast) gradient and a 40 min (slow) gradient, respectively. Additional peaks from protein impurities in the carbonic anhydrase sample are also shown. The presence of two of these impurities is consistent with previous reports, which identified these as ubiquitin18 and superoxide dismutase.14,18 There is an unknown impurity peak that elutes after carbonic anhydrase, indicated in Figure 2A,B as peak 6. There are also many smaller peaks due to other impurities in the commercial proteins. The chromatograms show that the model protein mixture was well resolved for both gradients, with excellent peak shapes and signal-to-noise ratios. There is little congestion of peaks, giving a very low baseline between most of the peaks. As expected, the slower gradient provides higher resolution, which is most noticeable in the crowded region in the vicinity of peaks 5 and 6. The reproducibility of the chromatograms is detailed in Figure S1 of the Supporting Information, showing that the RSD is no more than 0.34% for each peak.
Figure 2.
Chromatograms from LC–MS of proteins presented on three different vertical scales for (A) a fast gradient of 1% to 70% acetonitrile in 10 min and (B) a slow gradient of 20% to 70% acetonitrile over 40 min. Proteins: 1, ribonuclease A; 2, ubiquitin; 3, superoxide dismutase; 4, trypsin inhibitor; 5, carbonic anhydrase; 6, impurity (m = 19.554, 19.657, 19.570 Da). (C and D) Data (○) and Gaussian fits (—) for ubiquitin peaks of chromatograms in panels A and B, respectively. (E and F) Data (○) and Gaussian fits (—) for carbonic anhydrase peaks from panels A and B, respectively.
Figure 2C,D shows that the ubiquitin peaks fit well to Gaussians for both fast and slow gradients. This indicates unusual efficiency since protein peaks invariably tail in reversed-phase LC. The peak widths (2σ) are shown to be quite small, 1.55 and 3.8 s for the fast and slow gradients, respectively. The peak capacity, nc, can be calculated from the peak standard deviation. Equation 1 shows the peak capacity depends on gradient time (wg) and the peak width (4σ)
| 1 |
The peak capacities are calculated to be 195 and 315 for the 10 and 40 min gradients, respectively. These rival the best peak capacities reported for the case of UV detection, although it is difficult to compare directly because peak capacity in the latter case was calculated from the full width at half-maximum (fwhm).
| 2 |
Equation 2 is equal to eq 1 only for a Gaussian function. For tailing peaks, eq 2 overestimates peak capacity. Since the chromatograms corresponding to the peak capacities in the work of Fekete et al. were not shown and the chromatograms that were shown had significant tailing, a comparison cannot be made with certainty. Alternatively, a direct comparison can be made with the results of Roth et al. for LC–MS using porous shell particles, where the chromatograms were shown, and furthermore, the same protein was used: ubiquitin.18 Their ubiquitin peak is quite asymmetric: the ratio for the trailing to leading half-widths at 10% of the peak height is 2.0. Using the actual base width at 10% height, rather than eq 2, the peak capacity is 70 for the 10 min gradient. This is almost 3-fold lower than what we report here or the same gradient. Hence the slip flow capillary provides a significant advantage in peak capacity.
The protein peaks in the chromatograms of Figure 2 are fraught with impurities. These observations emphasize that even a sample of one protein can be a mixture that is difficult to fully resolve by reverse-phase LC. The peak that is widest and fits most poorly to a Gaussian is that of carbonic anhydrase. This protein has previously been reported to exhibit a shoulder on its leading side.14 Figure 2E,F shows, respectively, that the slip-flow column nearly resolves the shoulder for the fast gradient and gives baseline resolution for the slow gradient. For the fast gradient, the main peak nearly fits to a Gaussian, although there is fine structure indicating that more than one component is present. For the slow gradient, a new shoulder emerges and there is significant new fine structure, indicating that many components are present. The main peak has at least four Gaussians contributing to its shape. Three replicate chromatograms presented in Figure S2 of the Supporting Information establish that the fine structure is reproducible rather than originating from nanospray noise. One can conclude that the 2-fold wider peak of carbonic anhydrase in this work and earlier work is caused mostly by protein heterogeneity rather than column performance.
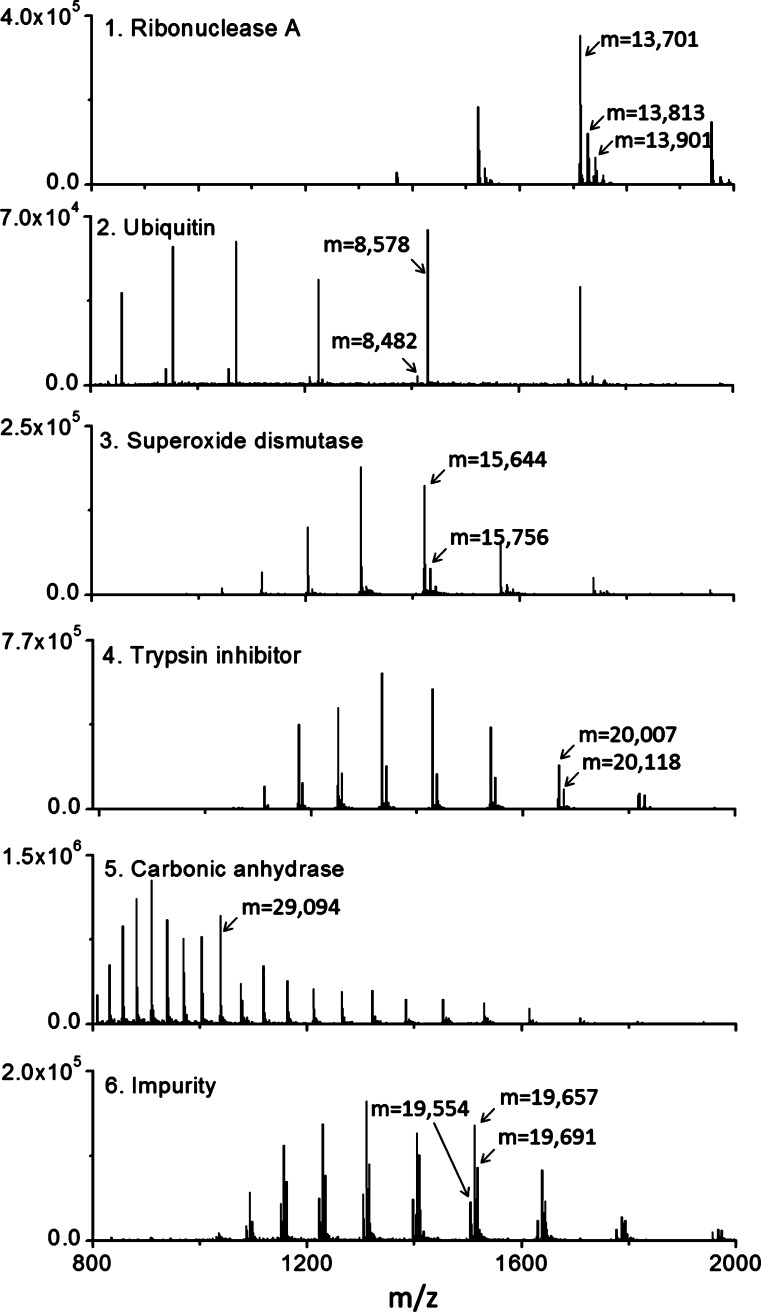
The mass spectra for all of the proteins studied in this work are presented in Figure 3, revealing multiple proteoforms that are resolvable with LTQ mass spectrometer used here. The peak for ribonuclease A is comparable in width to the peak for carbonic anhydrase, and the mass spectrum in the first panel of Figure 3 confirms the presence of four proteoforms. The masses of these peaks are denoted in the panel, and they differ by amounts suggestive of varying post-translational modification. Ubiquitin is the purest of the proteins, as proposed earlier on the basis of its Gaussian peak shape, and the mass spectrum in the second panel shows only one low-intensity proteoform in its mass spectrum. It is, of course, possible that there are other proteoforms that are not resolved by either the LTQ-MS or the chromatographic separation. The mass spectra for superoxide dismutase and trypsin inhibitor, given in panels 3 and 4, respectively, each show two major proteoforms. The mass spectrum for carbonic anhydrase in panel 5 shows that there are at least four proteoforms, and the mass spectrum for the impurity protein, given in panel 6, shows three proteoforms. Protein heterogeneity tends to give broader peaks and also complicates the determination of peak capacity. In top-down proteomics, multiple separation dimensions give complementary selectivity, such as charge, to help resolve the multiple proteoforms.
Figure 3.
Mass spectra for each of the peaks in the chromatogram of Figure 2A. The masses for the major proteoforms, calculated from the charge distributions, are indicated on each mass spectrum.
The slip flow capillary has the potential for delivering an order of magnitude higher sensitivity because the peaks are physically sharp, which translates to higher concentration. To illustrate, in Figure 2 the peak width (4σ) for ubiquitin is 3.1 s, and the volume follow rate is 200 nL/min; therefore, the injected moles are contained within a volume of only 10 nL. By contrast, for the similar separation by Roth et al.,18 the peak width (4σ) was 8.5 s, which is more than 2-fold longer in time units, and the volume flow rate was 5-fold higher, 1 000 nL/min; therefore, the injected moles were contained within a volume of more than 100 nL. The 10-fold smaller volume for the same number of moles injected gives the 10-fold higher sensitivity, and this is a consequence of the smaller physical width of the peak. Further, our column’s inner diameter was larger, so the calculated sensitivity factor is even greater. It is not possible to directly compare sensitivities because the choice of mass spectrometer and capillary tip diameter affect sensitivity. For a given system, the slip flow capillary will deliver more than a 10-fold higher concentration to the mass spectrometer due to the physically sharper peaks.
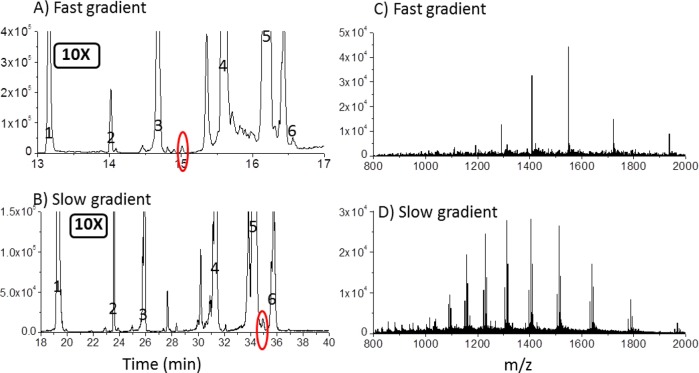
The high sensitivity and large peak capacity combine to give a wide dynamic range. This is illustrated in Figure 4, for which the same chromatograms in Figure 2 are plotted on 10× more sensitive scales. As an example, for the fast gradient, a small peak eluting at 15 min is circled in red in Figure 4A, and the mass spectrum for this peak is shown in Figure 4C. The S/N ratio is shown to be quite high, with the charge states from z = 8 to z = 13 are easily detected to reveal a mass of 15 485 ± 8 Da. This protein elutes between superoxide dismutase and trypsin inhibitor, and despite the fact that it is 50 to 100-fold lower in intensity than these two nearby peaks, its mass spectrum is easily interpreted. The mass spectra for the nearby small peaks are also easily interpreted (not shown). This illustrates that the fast gradient gives sharp peaks for high sensitivity while still having reasonable resolution to maintain a low baseline.
Figure 4.
Illustration of low background enabling small peaks to be characterized. (A and B) Same chromatograms as in Figure 2 but now plotted on a 10× expanded vertical scale. (C) Mass spectrum for the small peak in panel A that is circled in red, showing a S/N of 10 for this peak that is 100× smaller than the peak having maximum intensity. (D) Mass spectrum for the small peak in panel B that is circled in red, showing little contribution from the 100× larger carbonic anhydrase peak to which it is adjacent.
Slower gradients offer another advantage: resolving peaks where there is congestion in the chromatogram. In Figure 4B, for the slow gradient, a small peak eluting at 35 min is circled in red, and its mass spectrum is shown in Figure 4D. Despite the fact that this small peak is adjacent to the main carbonic anhydrase peak and it is only 1.8% of the height of the carbonic anhydrase peak, Figure 4D shows that there is little signal from carbonic anhydrase. The mass spectrum indicates that this peak is comprised of three proteoforms of an unknown protein, with mass differences suggestive of different post-translational modifications. The ability to detect such minority peaks is useful in conjunction with multidimensional separations, where the protein background would be low.
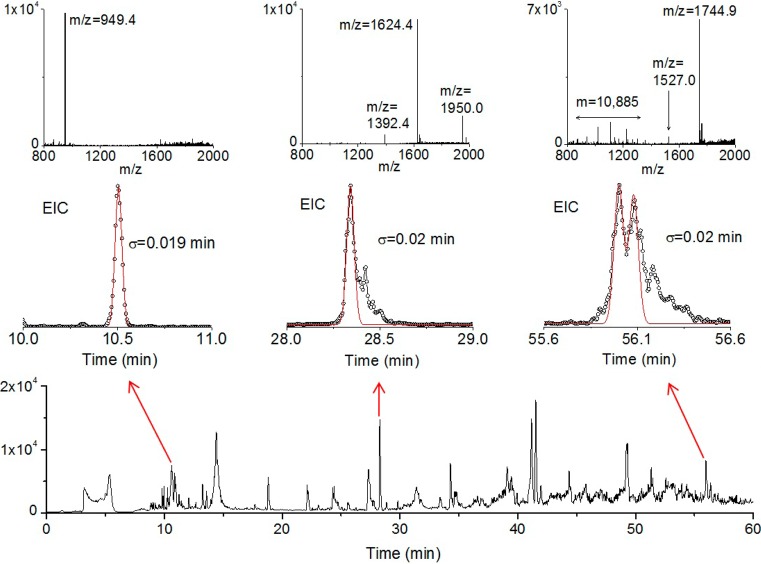
To investigate the application of the slip flow capillary to a very complex sample, the water-soluble fraction of an E. coli protein mixture was studied. Figure 5 shows the base-peak chromatogram for a 60 min gradient elution. The early eluting species that give sharp peaks in the vicinity of 9–12 min are peptides. The later eluting species are proteins, which give broader peaks due to multiple proteoforms. Three regions of the chromatogram were analyzed further by using extracted-ion chromatograms to find proteins that could reasonably be distinguished from the different proteoforms within the moderate resolving power of the LTQ mass spectrometer. The EIC for a peak eluting at 10.5 min is shown in Figure 5 to fit well to a Gaussian with a standard deviation 0.019 min. For the 60-min gradient, this width gives a computed peak capacity of 790. The mass spectrum shows a single peak m/z = 949.4, indicating that this is a peptide. The EIC for a peak eluting at 28.4 min showed a less homogeneous species, for which the tallest peak fit to a similar standard deviation, giving a computed peak capacity of 750. The mass spectrum indicates that this is from a protein of 9.7 kDa in mass. For elution times after 40 min, the base-peak chromatogram becomes quite congested because of the multiplicity of proteoforms for the higher molecular weight proteins. Among these, the EIC for a peak eluting at 56.0 min for a lower molecular weight species is shown in Figure 5 to have several overlapped peaks. The Gaussian fit gives an estimated peak width also of 0.02 min, again giving a computed peak capacity of 750. The mass spectrum shows this species either to be a peptide of mass 1744.9 Da or, more likely, a protein of mass 12.2 kDa. The results thus show that the efficiency is still high even in the presence of congestion for these late eluting proteins. A higher resolution mass spectrometer combined with this capillary would likely alleviate much of the congestion in the extracted-ion chromatograms. While the peaks are very sharp, in time units they are 2.6 s wide (fwhm), which is ample for a higher resolution mass spectrometer, such as the Orbitrap.
Figure 5.
Base-peak chromatogram of a cell lysate over a 60 min gradient is shown in the bottom panel. The arrows point to extracted-ion chromatograms (EIC) on expanded scales for three narrow peaks: early eluting (10.5 min), intermediate eluting (28.4 min), and late eluting (56.0 min). The standard deviations (σ) are shown on the EIC plots, and each is based on a Gaussian fit (red curve) to one or more peaks in the extracted-ion chromatogram. The mass spectrum is also shown for each of the three peaks, where the m/z with the highest signal was used in each case to generate the extracted-ion chromatogram. Gradient conditions: 20–55% acetonitrile over 60 min, with 0.5% formic acid and 0.02% TFA.
Conclusions
Slip flow imparts sufficient flow rates for submicrometer particles to be used for improving efficiency and sensitivity in reversed-phase LC–MS of intact proteins. Very high peak capacities are generated.
Acknowledgments
This work was supported by NIH under Grant R01-GM101464. We thank Professor W. Andy Tao of Purdue for use of the tip-puller.
Supporting Information Available
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): The senior author is part owner of a company that has licensed the intellectual property related to this technology.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- James P. Q. Rev. Biophys. 1997, 30, 279–331. [DOI] [PubMed] [Google Scholar]
- Capriotti A. L.; Cavaliere C.; Foglia P.; Samperi R.; Lagana A. J. Chromatogr., A 2011, 1218, 8760–8776. [DOI] [PubMed] [Google Scholar]
- Silberring J.; Ciborowski P. TrAC, Trends Anal. Chem. 2010, 29, 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. M.; Kelleher N. L. Nat. Methods 2013, 10, 186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLafferty F. W. Annu. Rev. Anal. Chem. 2011, 4, 1–22. [DOI] [PubMed] [Google Scholar]
- Shi S. D. H.; Hemling M. E.; Carr S. A.; Horn D. M.; Lindh I.; McLafferty F. W. Anal. Chem. 2001, 73, 19–22. [DOI] [PubMed] [Google Scholar]
- Tran J. C.; Zamdborg L.; Ahlf D. R.; Lee J. E.; Catherman A. D.; Durbin K. R.; Tipton J. D.; Vellaichamy A.; Kellie J. F.; Li M. X.; et al. Nature 2011, 480, 254–U141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveyra M. X.; Cuadrado-Corrales N.; Marcos A.; Barquero M. S.; Rabano A.; Calero M.; Saez-Valero J. J. Neurochem. 2006, 96, 97–104. [DOI] [PubMed] [Google Scholar]
- Rudrabhatla P.; Pant H. C. Topographic Regulation of Neuronal Intermediate Filament Proteins by Phosphorylation: In Health and Disease. In Cytoskeleton of the Nervous System; Nixon R. A., Yuan A., Eds.; Springer: New York, 2011; Vol. 3, pp 627–656. [Google Scholar]
- Zhang J.; Guy M. J.; Norman H. S.; Chen Y. C.; Xu Q. G.; Dong X. T.; Guner H.; Wang S. J.; Kohmoto T.; Young K. H.; et al. J. Proteome Res. 2011, 10, 4054–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson N. L.; Anderson N. G. Mol. Cell. Proteomics 2002, 1, 845–867. [DOI] [PubMed] [Google Scholar]
- Wang Y. J.; Balgley B. M.; Rudnick P. A.; Lee C. S. J. Chromatogr., A 2005, 1073, 35–41. [DOI] [PubMed] [Google Scholar]
- Everley R. A.; Croley T. R. J. Chromatogr., A 2008, 1192, 239–247. [DOI] [PubMed] [Google Scholar]
- Vellaichamy A.; Tran J. C.; Catherman A. D.; Lee J. E.; Kellie J. F.; Sweet S. M. M.; Zamdborg L.; Thomas P. M.; Ahlf D. R.; Durbin K. R.; et al. Anal. Chem. 2010, 82, 1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr J.; Swart R.; Samonig M.; Bohm G.; Huber C. G. Proteomics 2010, 10, 3598–3609. [DOI] [PubMed] [Google Scholar]
- Brice R. W.; Zhang X.; Colon L. A. J. Sep. Sci. 2009, 32, 2723–2731. [DOI] [PubMed] [Google Scholar]
- Fekete S.; Berky R.; Fekete J.; Veuthey J. L.; Guillarme D. J. Chromatogr., A 2012, 1236, 177–188. [DOI] [PubMed] [Google Scholar]
- Roth M. J.; Plymire D. A.; Chang A. N.; Kim J.; Maresh E. M.; Larson S. E.; Patrie S. M. Anal. Chem. 2011, 83, 9586–9592. [DOI] [PubMed] [Google Scholar]
- Eeltink S.; Wouters B.; Desmet G.; Ursem M.; Blinco D.; Kemp G. D.; Treumann A. J. Chromatogr., A 2011, 1218, 5504–5511. [DOI] [PubMed] [Google Scholar]
- Wei B. C.; Rogers B. J.; Wirth M. J. J. Am. Chem. Soc. 2012, 134, 10780–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin D. S.; Wei B. C.; Fogiel A. J.; Staats S. L.; Wirth M. J. Anal. Chem. 2010, 82, 2175–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns S.; Grinias J. P.; Blue L. E.; Jorgenson J. W.; Tallarek U. Anal. Chem. 2012, 84, 4496–4503. [DOI] [PubMed] [Google Scholar]
- Rogers B. J.; Birdsall R. E.; Wu Z.; Wirth M. J. Anal. Chem. 2013, 85, 6820–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B. C.; Malkin D. S.; Wirth M. J. Anal. Chem. 2010, 82, 10216–10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y.; Rappsilber J.; Andersen J. S.; Mann M. J. Chromatogr., A 2002, 979, 233–239. [DOI] [PubMed] [Google Scholar]
- Rogers B. J.; Wirth M. J. ACS Nano 2013, 7, 725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.