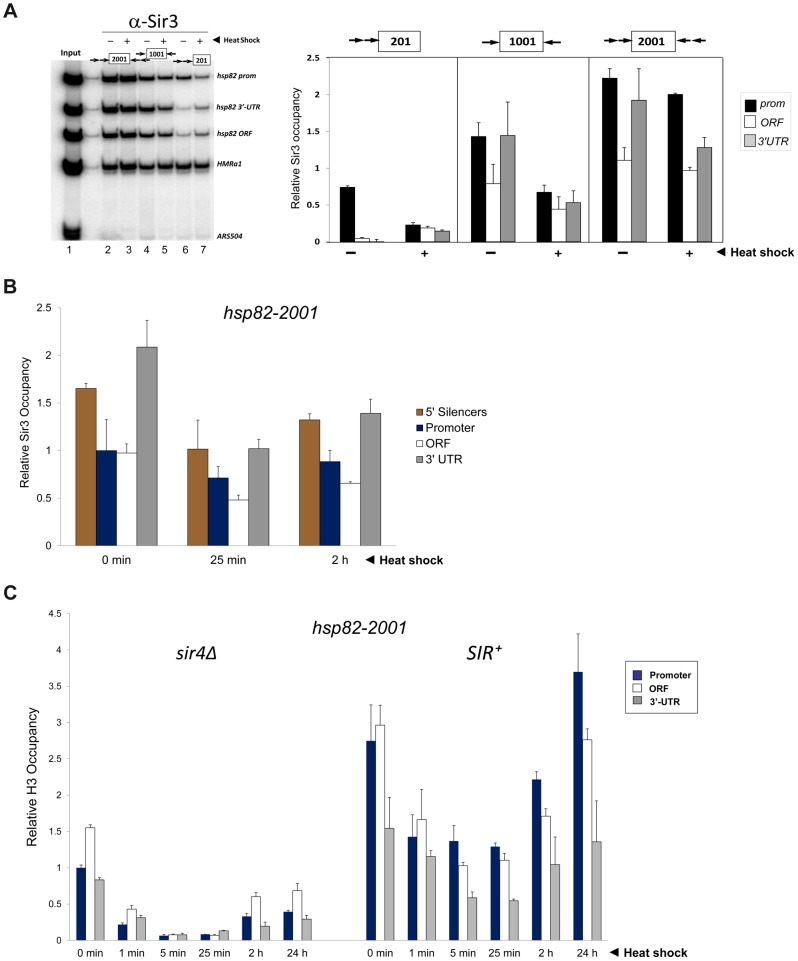
Figure 2. Retention of the Sir protein complex and increased nucleosome density and stability at the heat shock-induced hsp82-2001 transgene.
(a) In vivo crosslinking analysis of Sir3 at the promoter, ORF and 3′-UTR of hsp82-201, hsp82-1001 and hsp82-2001. Crosslinked chromatin, sheared to a mean size of ∼0.5 kb, was isolated from cells cultivated at 30°C and either maintained at that temperature or subjected to a 20 min 39°C heat shock (HS) (− and +, respectively). Following immunoprecipitation, crosslinks were reversed and purified DNA was subjected to quantitative multiplex PCR in the presence of [α-32P] dATP using primers specific for the five loci indicated. A gel analysis of multiplex PCR products is presented on the left, while a summary of three independent experiments (means ± S.E.) is presented on the right. Input sample (lane 1), derived from strain EAS2011, represents 4% of soluble chromatin used in the corresponding IP (lane 2). In the histogram, Sir3 occupancy at each hsp82 transgene was normalized to its occupancy at HMRa1. prom, promoter. (b) ChIP analysis of Sir3 at hsp82-2001 as in A, except that cells were subjected to the indicated heat shock time course and quantification of Sir3 abundance was performed using Real Time qPCR. Sir3 abundance at the indicated regions was normalized to its occupancy at HMRa2; illustrated are means ± S.D. (N = 2; qPCR = 4). (c) Histone H3 abundance at the promoter, ORF and 3′-UTR of hsp82-2001 in sir4Δ and SIR+ contexts as indicated, normalized to its occupancy at ARS504. Cultures were maintained at 30°C (0 min) or subjected to an instantaneous 39°C upshift for the times indicated. Quantification performed as in B; depicted are means ± S.D.; N = 2, qPCR = 4.