Figure 3. nsSNPs disrupting PDZ-binding motifs are only slightly under-represented in human proteins.
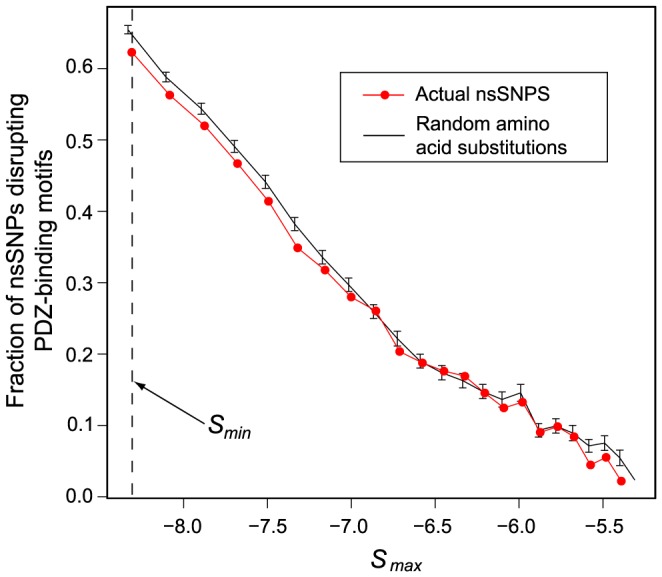
The red curve shows the fraction of known nsSNPs predicted to disrupt PDZ-binding motifs for different values of the threshold Smax. The black curve shows the same data for random amino acid substitutions on residues affected by nsSNPs in human C-termini (error bars show standard deviation for 1000 randomization of nsSNPs).
