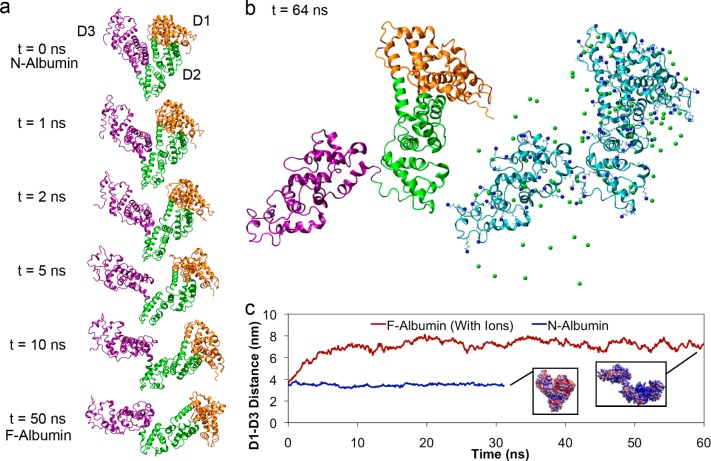
Figure 2.
Partial unfolding simulations of albumin with titratable residues set to pH 3.5 ionization states. Orange, green, and purple regions denote domains 1, 2, and 3 respectively. (a) Snapshots of albumin conformations simulation during partial electrostatically triggered denaturation. (b) Final simulation conformations of albumin at pH 3.5. Locations of positive charges and counterions are represented on the right. (c) Distance measured between the center of mass of domain 1 and domain 3 during simulations with counterions (red) in comparison with physiological albumin at pH 7.4 (blue). Insets depict albumin final conformations along each path and are colored with their electrostatic surface potential at the vdW distance (blue is positive, and red is negative).