Figure 4.
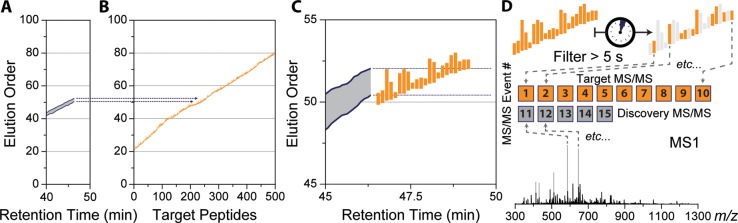
Following determination of the current elution order range (A), target peptides (B) sharing a similar elution order value are selected (C, rectangles represent individual peptides). Peptide targets within the elution order range are filtered based on when they were last sampled for MS/MS (D), leaving only targets that have been waiting the longest (e.g., > 5 s, highlighted rectangles). Those filtered peptides are then immediately sampled by MS/MS, regardless of MS1 detection (D). Unfilled MS/MS events are automatically filled with m/z features picked by the intensity-based DDA algorithm using normal sampling parameters (e.g., dynamic exclusion, intensity threshold, charge state exclusion, etc.).
