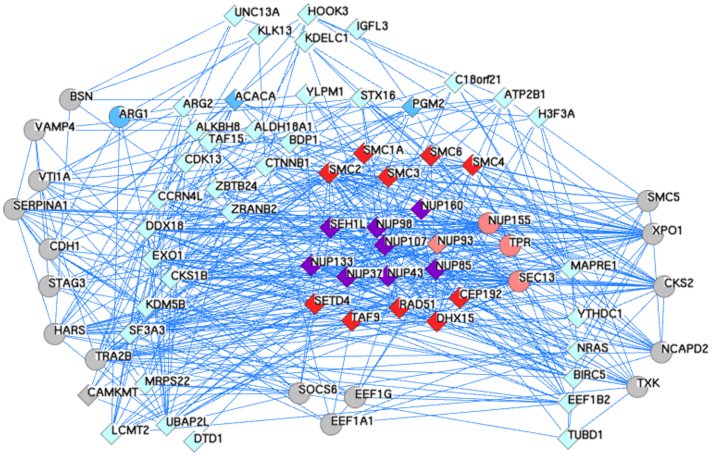
Figure 5. An expanded network of Y-Nup interactions from high-throughput experiments and the discovered multi-domain architectures.
Using as queries the human gene symbols of the molecules with coordinated gene expression patterns from the Y-Nup enriched middle block (see Figure 4), the resulting network is extracted by GeneMANIA with default parameters (only co-expression, physical and genetic interaction networks are retained). Queries were 60 (of which 3 are not found as interacting), depicted as diamonds (57 in total). An additional 20 genes are discovered by GeneMANIA, depicted as circles. The total genes in this network amount to 77, encompassing a number of known associations of Y-Nups (shown in purple, 8 in number) with other nucleoporins (shown in pink, 4 in number – including Sec13, not in query), e.g. TPR58 and related molecules (shown in light blue, 35 in number), e.g. EXO159. The reported molecules by GeneMANIA include interactions from large-scale experiments not further discussed (shown in grey, 17 in number). The three coordinated gene expression instances regarded as negatives in this study (shown in cyan, 3 in number) are ARG1 (curiously reported by GeneMANIA, thus shown as circle), ACACA and PGM2. The query molecule C2orf34 (synonym: CAMKMT, thus shown as diamond) is also reported by GeneMANIA. The six discovered novel domain associations (shown in purple, 10 in number), include the five of the six molecules with highest support (except CcmE/CycJ, Table 1) and five SMC paralogs (SMC1A, SMC2-4, SMC6), not previously found in association with Y-Nups. GeneMANIA reports no evidence for the association of NUP160-RAD51, NUP98-SET, NUP43-DHX15 and SEH1-TAF9, while providing strong evidence for SEH1-CEP19260,61,62. Common genes between PINA & GeneMANIA include other nucleoporins (e.g. NUP93) or others (e.g. EEF1G, Elongation factor 1-y) (see Methods). The annotated layout and GeneMANIA results with supporting literature are available in Data Supplement DS11.