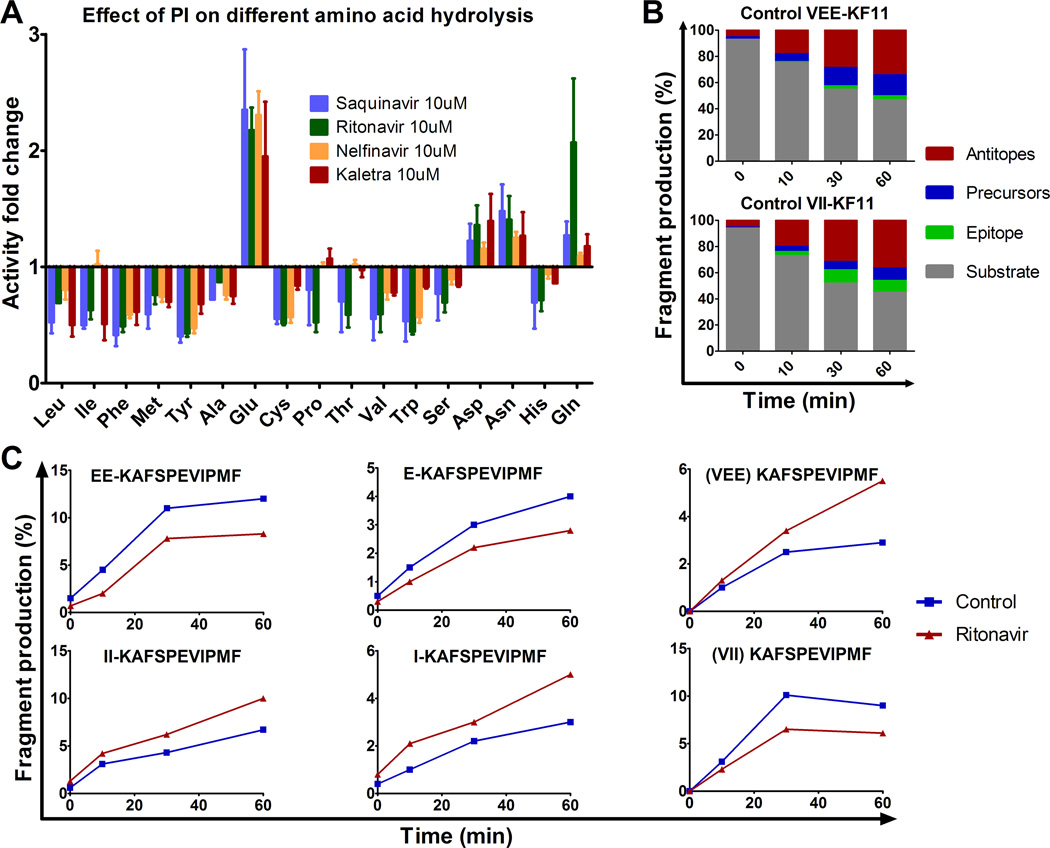
FIGURE 5.
HIV PIs modify HIV peptide degradation in a sequence-specific manner. (A) The hydrolysis of various amino acids was measured with fluorogenic substrate in PBMC pretreated with 10 µM Saquinavir (blue), Ritonavir (green), Nelfinavir (yellow) or Kaletra (red). Fluorescence emission was measured over 1 hour and the maximum slope of fluorescence was measured in each condition. The fold difference of the maximum slope of PI-treated PBMC over DMSO control was calculated for each condition. Data represent average results of 4 different experiments using 4 different PBMC donors. (B) The 3KF11 (VEEKAFSPEVIPMF) (upper panel) and 2I-3KF11 (VIIKAFSPEVIPMF) (lower panel) peptides were degraded using PBMC extracts. The resulting degradation products were analyzed by LC-MS-MS at 0, 10, 30 and 60 min. The distribution of substrate (3KF11 or 2I-3KF11, in grey), epitope (KF11, in green), precursors (blue) or antitopes (red) is shown at each time point. (C) The relative amount of epitope precursors (EEKAFSPEVIPMF and EKAFSPEVIPMF) and epitope KF11 (upper panels), and 2I-3KF11 epitope precursors (IIKAFSPEVIPMF and IKAFSPEVIPMF) and epitope (lower panels) were calculated at each time point of degradation with extracts pretreated with DMSO (control, blue) or with Ritonavir (red). Panels B and C are one experiment representative of two independent experiments using PBMC extracts from 2 different donors and run in duplicate on the mass spectrometer.