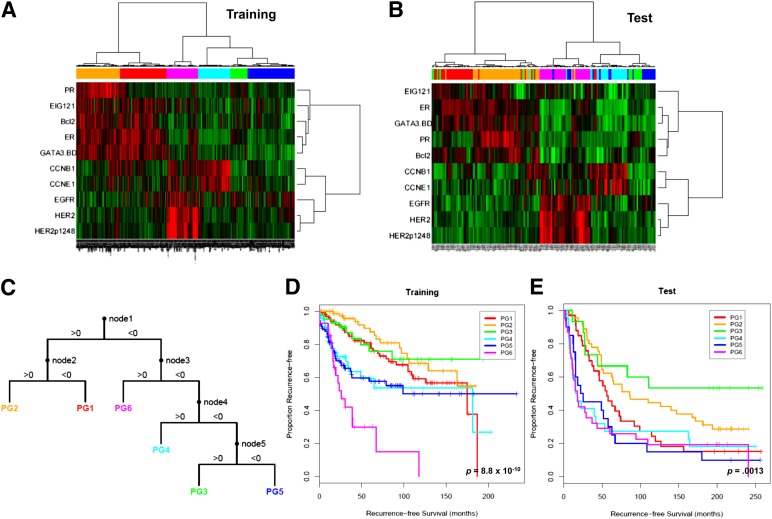
Figure 1.
Supervised clustering of breast cancers with quantification data for 10 proteins derived using reverse-phase protein arrays. The 712 breast tumor samples (training set) (A) were clustered with the 10 markers using an “uncentered correlation” distance metric along with the Ward linkage rule. This analysis yielded six subgroups (BG1–6). The 168 breast tumor samples (test set) (B) were subgrouped into one of six groups (PG1–6) using the decision tree (C) that was derived from the training set. Patients in the six subgroups differed significantly in their recurrence-free survival in both training (D) and test (E) sets. Reprinted from Gonzalez-Angulo AM, Hennessy BT, Meric-Bernstam F et al. Functional proteomics can define prognosis and predict pathologic complete response in patients with breast cancer. Clinical Proteomics, 2011;8:11.