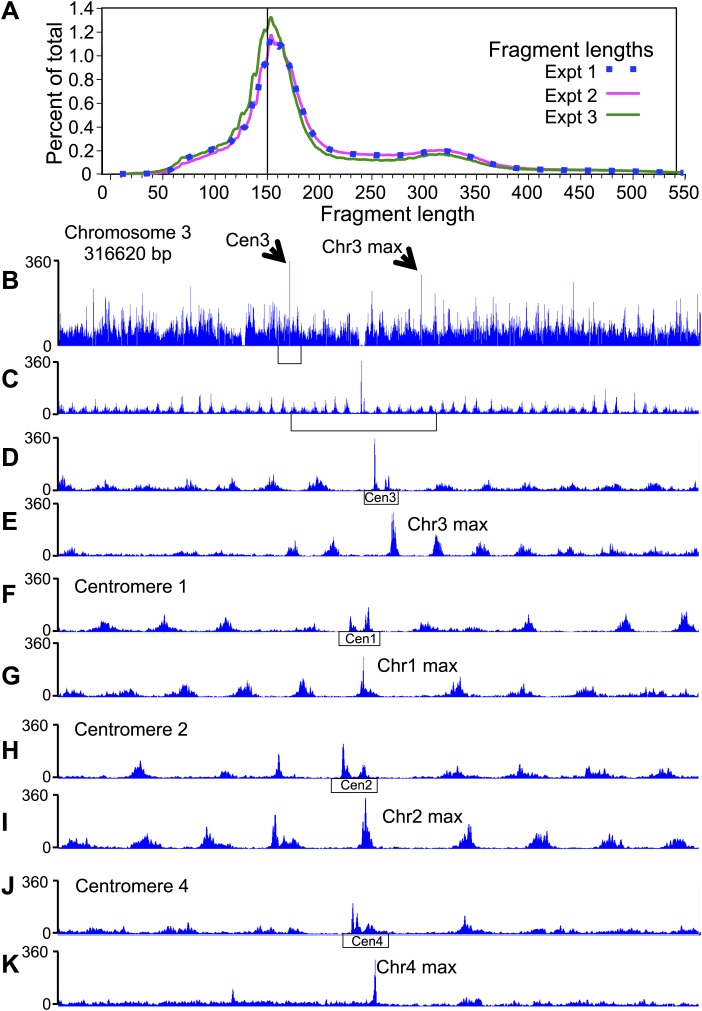
Figure 2. Pairs of cleavage clusters occupy CDEs.
(A) Fragment length distributions for three experiments. For each length in base pairs, the percentage of the total number of mapped fragments is plotted. Experiment 1 (using Phusion DNA polymerase): 27,871,803 fragments; Experiment 2 (using KAPA polymerase): 62,926,977 fragments; Experiment 3: 81,473,515 fragments (biological replicate using KAPA). (B–K) Examples of cleavage profiles around centromeres, where tracks represent the total number of fragment ends within successive 10-bp windows. (B) Chromosome 3 profile, where the position of the centromere and of the most frequently cleaved nucleotide position on the chromosome are indicated. (C) Expansion of the region indicated by the bracket in (B). (D) Expansion of the region indicated by the bracket in (C), where the extent of Cen3 is indicated. (E) Expansion of the region around the most frequently cleaved nucleotide position at the same scale as in (D). (F) and (G) Same as (D) and (E), respectively, for Chromosome 1. (H) and (I) Same as (D) and (E), respectively, for Chromosome 2. (J) and (K) Same as (D) and (E), respectively, for Chromosome 4. (See also Figure 2—figure supplement 1).