Table 3.
Scaffold, compound, potency (EC50 by ELISA), and calculated physicochemical parametersa for the 32 confirmed PrPC SPC hits in T98G cells.
| Scaffold | Compound | # | Structure | MW | EC50 ± SEM (μM)b | n | TPSA | xlogP | HBA/HBD |
|---|---|---|---|---|---|---|---|---|---|
| Amide | IND82865 | 1 |
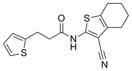
|
316.4 | 0.15 ± 0.04 | 4 | 52.9 | 4.54 | 2/1 |
| IND84911 | 2 |
|
342.1 | 0.45 ± 0.10 | 6 | 42.0 | 3.22 | 2/1 | |
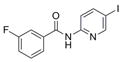
| IND116063 | 3 |
|
316.7 | 0.65 ± 0.06 | 4 | 58.6 | 2.88 | 3/1 | |
| IND85671 | 4 |
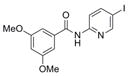
|
384.2 | 1.31 ± 0.35 | 7 | 60.4 | 3.04 | 4/1 | |
| IND85692 | 5 |
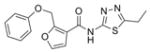
|
329.4 | 2.24 ± 0.39 | 6 | 77.6 | 4.44 | 5/1 | |
| IND87406 | 6 |
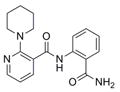
|
324.4 | 1.02 ± 0.09 | 18 | 88.3 | 1.98 | 3/1 | |
| IND116065 | 7 |
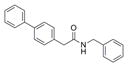
|
301.4 | 0.44 ± 0.11 | 6 | 29.1 | 5.05 | 1/1 | |
|
| |||||||||
| Aminothiazole | IND126306 | 8 |
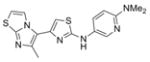
|
356.5 | 0.22 ± 0.06 | 4 | 58.3 | 4.28 | 5/1 |
| IND126328 | 9 |
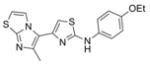
|
356.5 | 0.03 ± 0.01 | 4 | 51.4 | 5.59 | 4/1 | |
| IND9756 | 10 |
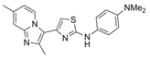
|
363.5 | 0.11 ± 0.01 | 3 | 45.5 | 5.29 | 4/1 | |
| IND115948 | 11 |
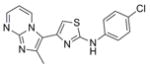
|
341.8 | 2.44 ± 1.37 | 6 | 55.1 | 4.49 | 4/1 | |
|
| |||||||||
| Chromene | IND8541 | 12 |
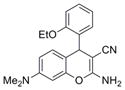
|
335.4 | 0.14 ± 0.02 | 4 | 71.5 | 3.37 | 5/1 |
| IND17990 | 13 |
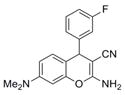
|
309.3 | 0.03 ± 0.00 | 6 | 62.3 | 3.1 | 4/1 | |
| IND126322 | 14 |
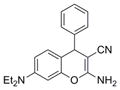
|
319.4 | 0.17 ± 0.01 | 4 | 62.3 | 3.82 | 4/1 | |
| IND126361 | 15 |
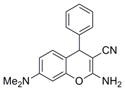
|
291.4 | 0.12 ± 0.02 | 3 | 62.3 | 3 | 4/1 | |
| IND23846 | 16 |
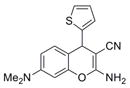
|
297.4 | 0.22 ± 0.01 | 6 | 62.3 | 2.95 | 4/1 | |
|
| |||||||||
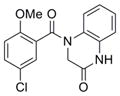
| Fused pyrrole | IND73602 | 17 |
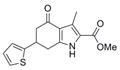
|
289.4 | 0.41 ± 0.10 | 4 | 59.2 | 3.09 | 4/1 |
| IND116088 | 18 |
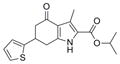
|
317.4 | 0.24 ± 0.07 | 3 | 59.2 | 4.01 | 4/1 | |
| IND126429 | 19 |
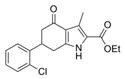
|
331.8 | 0.45 ± 0.03 | 4 | 59.2 | 4.18 | 4/1 | |
| IND126432 | 20 |
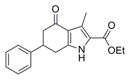
|
297.4 | 0.11 ± 0.02 | 4 | 59.2 | 3.56 | 4/1 | |
| IND87564 | 21 |
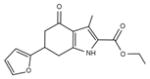
|
287.3 | 0.68 ± 0.34 | 6 | 83.96 | 2.80 | 4/1 | |
|
| |||||||||
| Piperazine | IND30802 | 22 |
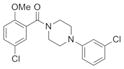
|
365.3 | 0.17 ± 0.02 | 3 | 32.8 | 4.17 | 4/0 |
| IND116050 | 23 |
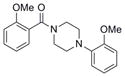
|
326.4 | 3.24 ± 2.35 | 3 | 42 | 2.89 | 4/0 | |
| IND126339 | 24 |
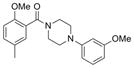
|
340.4 | 0.27 ± 0.02 | 4 | 42 | 3.17 | 4/0 | |
|
| |||||||||
| Sulfonamide | IND5418 | 25 |
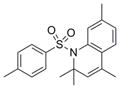
|
341.5 | 3.77 ± 3.11 | 3 | 37.4 | 4.87 | 2/0 |
| IND16365 | 26 |
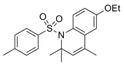
|
371.5 | 0.69 ± 0.17 | 3 | 46.6 | 4.96 | 3/0 | |
| IND18762 | 27 |
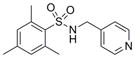
|
290.4 | 4.13 ± 0.66 | 4 | 59.1 | 2.11 | 3/1 | |
| IND86287 | 28 |
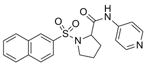
|
381.5 | 1.52 ± 0.66 | 4 | 79.4 | 2.35 | 4/1 | |
| IND116071 | 29 |
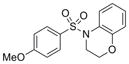
|
305.4 | 1.32 ± 0.32 | 4 | 55.8 | 2.15 | 4/0 | |
| IND126331 | 30 |
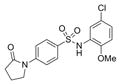
|
380.9 | 0.51 ± 0.17 | 3 | 75.7 | 3.33 | 4/1 | |
|
| |||||||||
| Bi-aryl ketone | IND1323 | 31 |
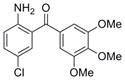
|
321.8 | 0.09 ± 0.01 | 4 | 70.8 | 3.21 | 5/1 |
| IND5672 | 32 |
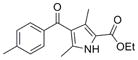
|
285.3 | 0.53 ± 0.04 | 6 | 59.2 | 3.68 | 3/1 | |
Physicochemical parameters included MW = molecular weight; TPSA = topological polar surface area relating to N and O atoms (TPSA_NO); xlogP = lipophilicity coefficient; HBA = H-bond acceptor; and HBD = H-bond donor, calculated in Vortex v2011, Dotmatics Limited.
Mean ELISA EC50 values based on n ≥ 3. Twenty-nine compounds were also tested in N2a-cl3 cells, three of which showed good potency, noted in parentheses.
