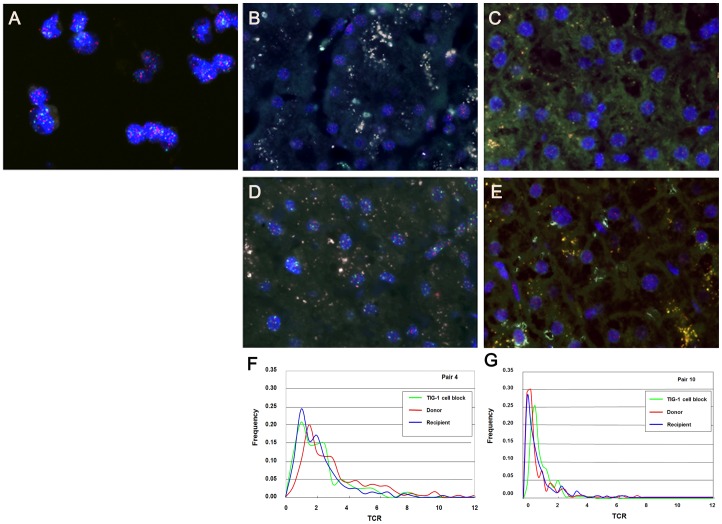
Figure 1. Representative Q-FISH images of TIG-1 and paired liver tissues (red: Cy3, telomere signals; green: FITC, centromere signals; blue: DAPI, nuclei).
(A) Q-FISH image reveals the TIG-1 cell block placed on the same slide together with liver sections. Telomere (Cy3, red) and centromere (FITC, green) signals are evident (original magnification ×400). (B) Q-FISH images of pair 4 donor. Telomere and centromere signals are evident in the nuclei (original magnification ×400). (C) Q-FISH images of pair 10 donor. (original magnification ×400). (D) Q-FISH image of pair 4 recipient in the lower NTCR group reveals weaker telomere signals (red) than those in the paired donor (Figure 1B) (original magnification ×400). (E) Q-FISH images of pair 10 recipient, showing brighter telomere signals (red) than those in the paired donor (Figure 2D) (original magnification ×400). (F) Distributions of the telomere intensity given by telomere-to-centromere ratio (TCR) in TIG-1 and hepatocytes from paired liver tissues of pair 4 samples. Green: TIG-1 cells in a cell block, red: hepatocytes in donor, blue: hepatocytes in recipient grafted liver. (G) TCR distribution in TIG-1, and hepatocytes from paired liver tissues of pair 10 samples.