Abstract
We introduce a simple MODelability Index (MODI) that estimates the feasibility of obtaining predictive QSAR models (Correct Classification Rate above 0.7) for a binary dataset of bioactive compounds. MODI is defined as an activity class-weighted ratio of the number of the nearest neighbor pairs of compounds with the same activity class versus the total number of pairs. The MODI values were calculated for more than 100 datasets and the threshold of 0.65 was found to separate non-modelable from the modelable datasets.
Cheminformatics approaches such as QSAR modeling are applied widely for the analysis of growing collections of bioactive compounds in private and publicly-available online repositories such as ChEMBL1 and PubChem2. The resulting models are used for designing new bioactive molecules or identifying those by virtual screening; thus, it is imperative that such models have reliable external predictive power.
We3,4 and others5 have shown previously that the predictivity of QSAR models is directly influenced by various dataset characteristics (e.g., size, chemical diversity, activity distribution, presence of activity cliffs, etc.) as well as the modeling workflow (e.g., dataset curation, variable selection, external validation, consensus modeling, use of applicability domain, etc.) utilized to build, select, and validate the models.6 It is not uncommon for cheminformaticians to employ many different descriptor types, machine-learning techniques, validation workflows, etc., in a combinatorial manner in order to maximize the prediction performance of QSAR models7. Such attempts are time and resource consuming, especially when the datasets contain more than a few thousands compounds (which becomes more and more common). However, extensive investigations of large collections of datasets suggests that it is often impossible to build models with appreciable external predictive power even when the most sophisticated algorithms and rigorous modeling workflows are employed.8
Herein, we introduce a concept of “dataset modelability”, i.e., an a priori estimate of the feasibility to obtain externally predictive QSAR models for a dataset of bioactive compounds. This concept has emerged from analyzing the effect of so called “activity cliffs” on the overall performance of QSAR models. Indeed, in a seminal observation, Maggiora9 suggested that the presence of “activity cliffs”, i.e., very similar compounds with very different activities, present significant challenges for QSAR modeling. Thus, SALI10 and ISAC11 scores were developed for identifying activity cliffs based on ligand- and structure-based approaches, respectively. A recent excellent review from the Bajorath’s group12 discusses many issues posed by the activity cliffs for cheminformatics investigations.
The effect of activity cliffs in a dataset on the process and outcome of the QSAR modeling can be illustrated by the case of stereoisomers. Indeed, the nature and the actual number of activity cliffs not only depends on the endpoint and overall data quality, but also on the choice of descriptors used to characterize chemical structures. When stereoisomers (as well as some other types of isomers) are present in a dataset it is important to explore whether descriptors used to characterize compounds are sensitive to chirality. Obviously, when using two-dimensional (2D) descriptors, any pair of stereoisomers appears as duplicates. In this case, prior to the actual modeling of the chemical dataset, one should carefully check the experimental properties reported for all the pairs of stereoisomers present in the set. If the target property values for stereoisomers are significantly different we have an extreme case of activity cliffs when two formally identical compounds have different activities and we have no ground to chose one over another; therefore such pairs should be removed from the dataset prior to model building (or 3D descriptors should be employed). On the other hand, if stereoisomers have similar activities, one of them could be kept for model development.
The obvious attention given to the problem of activity cliffs notwithstanding, to the best of our knowledge there has not been any exhaustive study to explore (i) how the number of activity cliffs in a given dataset correlates with the overall prediction performance of QSAR models for this dataset, (ii) whether such correlation is conserved across different datasets, and (iii) whether one could use the fraction of activity cliffs in a datasets to assess the overall possibility of success or failure for QSAR modeling. To this end, we propose a “MODelability Index” (MODI) as a quantitative means to quickly assess whether predictive QSAR model(s) can be obtained for a given chemical dataset. The current version of MODI is only applicable to binary endpoints but its extension to datasets of compounds with real activity values is also possible.
The MODI is computed based on the following considerations. For every compound in a dataset, we determine whether its first nearest neighbor, i.e., a compound with the smallest Euclidean distance from a given compound estimated in the entire descriptor space, belongs to the same or different activity class. In the latter case, the pair can be formally designated as an activity cliff. The number of nearest neighbor pairs that are not activity cliffs is counted for each class of compounds and is used to calculate MODI as follows:
| (Equation 1) |
where K is the number of classes (K=2 for binary datasets), Nisame is the number of compounds of i-th activity class that have their first nearest neighbors belonging to the same activity class i; Nitotal is the total number of compounds belonging to the class i.
The predictive performance of QSAR models is expressed as the correct classification rate (or balanced accuracy)13 calculated with 5-fold external cross-validation (QSAR_CCR). Note that QSAR_CCR could be also estimated from a formula similar to Equation 1, where Nisame would be the number of correctly predicted compounds belonging to i-th activity class. In general, we consider the QSAR model to have an acceptable predictive power if it affords QSAR_CCR equal or higher than 0.7 (see ref.6).
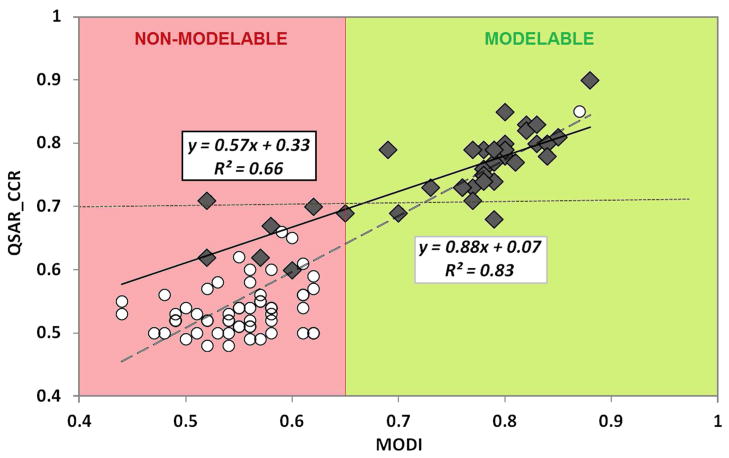
The utility of MODI was assessed initially using 42 diverse datasets related to pharmaceutical targets: MDR114, MDR1i14, six types of C. Elegans toxicity15, and 34 GPCR datasets16. All the details related to QSAR modeling including molecular descriptors, machine learning techniques, and the results of the modeling are given in the Supplementary Materials. Prior to the analysis, all datasets considered in this study were rigorously curated according to the workflow developed in our laboratory3. QSAR models were built using Dragon17 and, for a few cases, MOE18 descriptors, and one or several machine learning techniques including k-nearest neighbors QSAR (kNN), Support Vector Machines (SVM), and Random Forest (RF). When examining the results, we have found a significant correlation (R2=0.66) between MODI and models’ predictivity (i.e., QSAR_CCR) as illustrated in Figure 1. Although this correlation is not high enough to predict the exact QSAR_CCR value from MODI, it still affords a reasonable assessment whether the dataset is modelable or not. Obviously, this initial collection had a bias towards modelable datasets (QSAR_CCR > 0.7) because these datasets included compounds tested in high-quality in vitro assays against specific molecular targets.
Figure 1.
Correlation between QSAR_CCR (Y-axis) and MODI (X-axis) for 42 miscellaneous (black diamonds) and 60 ToxCast datasets (hollow circles). Regression lines and the corresponding equations are shown for 42 datasets (solid line) and all 102 datasets (dashed line).
Recently, Thomas et al.8 published the results of massive QSAR calculations for ToxCast (www.epa.gov/ncct/toxcast/) datasets with the goal of predicting 60 ToxRefDB (epa.gov/ncct/toxrefdb/) in vivo toxicity endpoints. They used both chemical descriptors and the results of in vitro assays considered as independent variables as well as a combination of chemical and biological descriptors to predict in vivo toxicities but for this study we only used QSAR models built with conventional chemical descriptors (see Supplemental Materials for details). The authors8 employed all possible combinations (as many as 84) of descriptors, modeling techniques, and rigorous validation workflows. However, no models with significant predictive power (much greater than 50% for binary classification models) were obtained with only one exception. Thus, we enriched our initial pool of datasets with those taken directly from reference8. Similarly to the initial pool of 42 sets, we chose models with the highest QSAR_CCR values among the 84 models obtained by Thomas et al.8 for each of the 60 in vivo endpoints (see Figure 1).
As Thomas et al.8 have already established that no good models have been generated for those datasets, we should have expected low MODI values. Indeed, we have found that 59 out of 60 datasets (represented as a cluster of white circles in the lower left part of Figure 1) were characterized by low MODI values, in full agreement with the failure of Thomas et al.8 to develop QSAR models with significant predictive power. The only exception was the rat cholinesterase inhibition dataset for which MODI = 0.83 and QSAR_CCR = 0.82 (white circle in upper-right part of Figure 1). Similar results have been generated using models built with biological descriptors (data not shown). Interestingly, the authors commented on their findings by positing that in vitro assays have “limited applicability for predicting in vivo chemical hazards using standard statistical classification methods”, i.e., questioning the in vitro to in vivo extrapolation paradigm as applied to the Toxcast datasets. On the contrary, our studies suggest that the datasets employed in that study, with one exception, were merely not amenable to the development of predictive models because of a large fraction of activity cliffs.
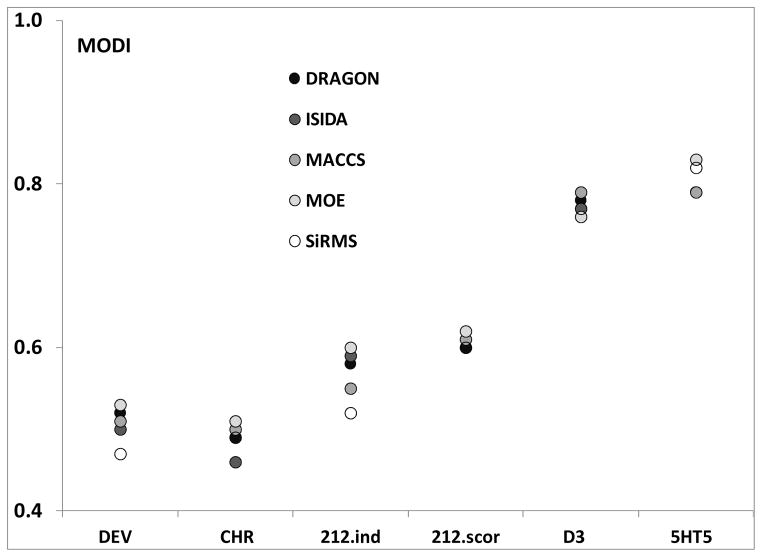
In order to study how the choice of chemical descriptors influences the MODI values, we computed different types of descriptors (SiRMS19, Dragon17, ISIDA20, MACC21, and MOE18) for six different datasets: DEV8 (n= 241 compounds), CHR8 (n= 238), 212.ind15 and 212.scor15 (the same 212 compounds but different endpoints), D316 (n= 1509), and 5HT516 (n= 195). As shown in Figure 2, the types of chemical descriptors had rather weak influence on MODI. Additional details about these datasets can be found in Supplementary Materials.
Figure 2.
Low variability of MODI when different types of chemical descriptors are used.
Overall, for all 102 datasets (42 pharmaceutical targets plus 60 Toxcast datasets), the correlation between QSAR_CCR and MODI values was high (R2=0.83) demonstrating the validity of MODI as a reliable simple metric to evaluate the dataset modelability a priori. The correlation shown in Figure 1 affords a simple means to estimate the highest QSAR_CCR value from that of MODI. However, as a possible pitfall, this correlation may have limited generality: for instance, only a small number of datasets had MODI values ranging between 0.65 and 0.75. Thus, additional studies with more datasets are needed to validate the quantitative relationship between MODI and the best QSAR_CCR.
In summary, we have introduced the QSAR MODI as a simple metric for rapidly assessing whether a given chemical dataset is likely to be modelable or not. The results of this study suggest (cf. Figure 1) that a MODI value for a given dataset below 0.65 indicates that one should not expect to achieve QSAR models with significant predictive power, whereas MODI>0.65 implies that the underlying dataset is modelable and will have QSAR_CCR greater than 0.7. As follows from Figure 1, there are very few outliers from this general simple rule.
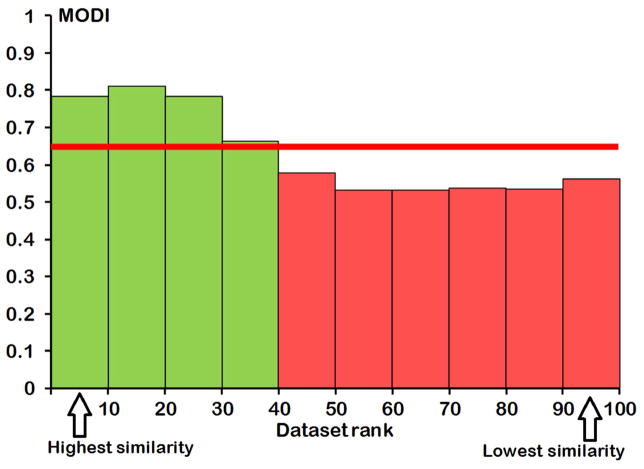
This study begs a natural question as to why some datasets are modelable whereas others are not. As follows from the simple formula for MODI (see Eq. 1), this index depends on the fraction of activity cliffs in a dataset. Activity cliffs are not uncommon and indeed there are many examples of compound pairs that are highly similar to each other and yet have significantly different or opposite (in case of binary classification) activities12: these cases represent true activity cliffs. However, we shall point out that Eq. 1 does not require that the pairs of compounds defined formally as activity cliffs should be highly similar to each other. These pairs are defined as nearest neighbors only within a given dataset and they may not be highly similar to each other based on absolute similarity metric such as Tanimoto coefficient (Tc). The use of Eq. 1 to estimate MODI is based on a reasonable expectation (which is the foundation of the Active Analog principle widely used by both experimental and computational medicinal chemists) that similar compounds are expected to have similar activities; thus a “modelable” dataset is expected to have a large fraction of compound pairs that follow the Active Analog principle. On the other hand, many modern datasets especially relatively large ones evaluated for some general biological effect such as toxicity (e.g., Toxcast dataset explored by Thomas et al.8) may include rather diverse collections of chemicals that may exert the underlying biological effects through multiple mechanisms. In such cases, one should not have any rational expectation that two compounds with different chemical structure that appear as formal nearest neighbors should have similar end point effects, and in fact this is often not the case. It then follows that chemically diverse datasets tested for the same endpoint activity should contain a large fraction of activity cliffs making them non-modelable. Indeed, the analysis of datasets explored in this study suggests that as a rule, non-modelable datasets with relatively low MODI values also have relatively low average Tc values for all pairs of nearest neighbors and, vice versa, modelable datasets have relatively high Tc values (Fig. 3). It also suggests that, although a dataset with low MODI is not modelable as a whole, it may still contain subsets of compounds with high MODI for which local QSAR models can be built.
Figure 3.
MODI (Y-axis) vs. dataset rank (ordered by descending average structural similarity (Tc) between all pairs of nearest neighbors within a dataset; X-axis). Horizontal line at MODI=0.65 is a cut-off value separating modelable (green bars) vs non-modelable (red bars) datasets.
In conclusion, we suggest that MODI is a simple characteristic that can be easily computed for any dataset at the onset of any QSAR investigation. We hope that this simplicity will prompt our colleagues to compute and report MODI for any dataset they consider developing QSAR models for, which will enable further evaluation of the dataset modelability concept introduced in this study. Finally, we shall point out that studies reported herein for binary datasets can be easily extended for additional datasets with multi-class and continuous value activities.
Supplementary Material
Acknowledgments
This study was supported in part by NIH (grants GM66940 and GM096967) and EPA (grant RD 83499901). The authors thank Dr. Rusty Thomas for sharing datasets and statistical results of the analysis of the Toxcast datasets and Dr. Alex Sedykh for his interest to this study and fruitful discussions.
Footnotes
The authors declare no competing financial interest.
References
- 1. [accessed Mar 13, 2013];ChEMBL Database. https://www.ebi.ac.uk/chembl/
- 2. [accessed Oct 1, 2013];PubChem. http://pubchem.ncbi.nlm.nih.gov/
- 3.Fourches D, Muratov E, Tropsha A. Trust, but Verify: On the Importance of Chemical Structure Curation in Cheminformatics and QSAR Modeling Research. J Chem Inf Model. 2010;50:1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fourches D, Tropsha A. Using Graph Indices for the Analysis and Comparison of Chemical Datasets. Mol Inform. 2013;32:827–842. doi: 10.1002/minf.201300076. [DOI] [PubMed] [Google Scholar]
- 5.Young D, Martin D, Venkatapathy R, Harten P. Are the Chemical Structures in Your QSAR Correct? QSAR Comb Sci. 2008;27:1337–1345. [Google Scholar]
- 6.Tropsha A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol Inform. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 7.Zhu H, Tropsha A, Fourches D, Varnek A, Papa E, Gramatica P, Oberg T, Dao P, Cherkasov A, Tetko I. V Combinatorial QSAR Modeling of Chemical Toxicants Tested Against Tetrahymena Pyriformis. J Chem Inf Model. 2008;48:766–784. doi: 10.1021/ci700443v. [DOI] [PubMed] [Google Scholar]
- 8.Thomas RS, Black MB, Li L, Healy E, Chu TM, Bao W, Andersen ME, Wolfinger RD. A Comprehensive Statistical Analysis of Predicting in Vivo Hazard Using High-Throughput in Vitro Screening. Toxicol Sci. 2012;128:398–417. doi: 10.1093/toxsci/kfs159. [DOI] [PubMed] [Google Scholar]
- 9.Maggiora GM. On Outliers and Activity Cliffs--Why QSAR Often Disappoints. J Chem Inf Model. 2006;46:1535. doi: 10.1021/ci060117s. [DOI] [PubMed] [Google Scholar]
- 10.Guha R, Van Drie JH. Structure--Activity Landscape Index: Identifying and Quantifying Activity Cliffs. J Chem Inf Model. 2008;48:646–658. doi: 10.1021/ci7004093. [DOI] [PubMed] [Google Scholar]
- 11.Seebeck B, Wagener M, Rarey M. From Activity Cliffs to Target-Specific Scoring Models and Pharmacophore Hypotheses. Chem Med Chem. 2011;6:1630–1639. doi: 10.1002/cmdc.201100179. [DOI] [PubMed] [Google Scholar]
- 12.Stumpfe D, Hu Y, Dimova D, Bajorath J. Recent Progress in Understanding Activity Cliffs and Their Utility in Medicinal Chemistry. J Med Chem. 2013 doi: 10.1021/jm401120g. on-line early access. [DOI] [PubMed] [Google Scholar]
- 13.De Cerqueira Lima P, Golbraikh A, Oloff S, Xiao Y, Tropsha A. Combinatorial QSAR Modeling of P-Glycoprotein Substrates. J Chem Inf Model. 2006;46:1245–1254. doi: 10.1021/ci0504317. [DOI] [PubMed] [Google Scholar]
- 14.Sedykh A, Fourches D, Duan J, Hucke O, Garneau M, Zhu H, Bonneau P, Tropsha A. Human Intestinal Transporter Database: QSAR Modeling and Virtual Profiling of Drug Uptake, Efflux and Interactions. Pharm Res. 2013;30:996–1007. doi: 10.1007/s11095-012-0935-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyd WA, McBride SJ, Rice JR, Snyder DW, Freedman JH. A High-Throughput Method for Assessing Chemical Toxicity Using a Caenorhabditis Elegans Reproduction Assay. Toxicol Appl Pharmacol. 2010;245:153–9. doi: 10.1016/j.taap.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao G, Fourches D, Muratov E, Tropsha A. The QSARome of GPCRome. In Preparation. [Google Scholar]
- 17.Todeschini R, Consonni V. In: Handbook of Molecular Descriptors. Todeschini R, Consonni V, editors. Vol. 11. Wiley-VCH Verlag GmbH; Weinheim, Germany: 2000. p. 667. [Google Scholar]
- 18.Chemical Computing Group MOE. [accessed Oct 1, 2013]; http://www.chemcomp.com/index.htm.
- 19.Kuz’min VE, Artemenko AG, Muratov EN. Hierarchical QSAR Technology Based on the Simplex Representation of Molecular Structure. J Comput Aided Mol Des. 2008;22:403–421. doi: 10.1007/s10822-008-9179-6. [DOI] [PubMed] [Google Scholar]
- 20.Varnek A, Fourches D, Horvath D, Klimchuk O, Gaudin C, Vayer P, Solov’ev V, Hoonakker F, Tetko I, Marcou G. ISIDA-Platform for Virtual Screening Based on Fragment and Pharmacophoric Descriptors. Curr Comput Aided Drug Des. 2008;4:191–198. [Google Scholar]
- 21.Gunner FO, Hughes WD, Dumont ML. An Integrated Approach to Three-Dimensional Information Management with MACCS-3D. J Chem Inf Comput Sci. 1991;31:408–414. doi: 10.1021/ci00003a007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.