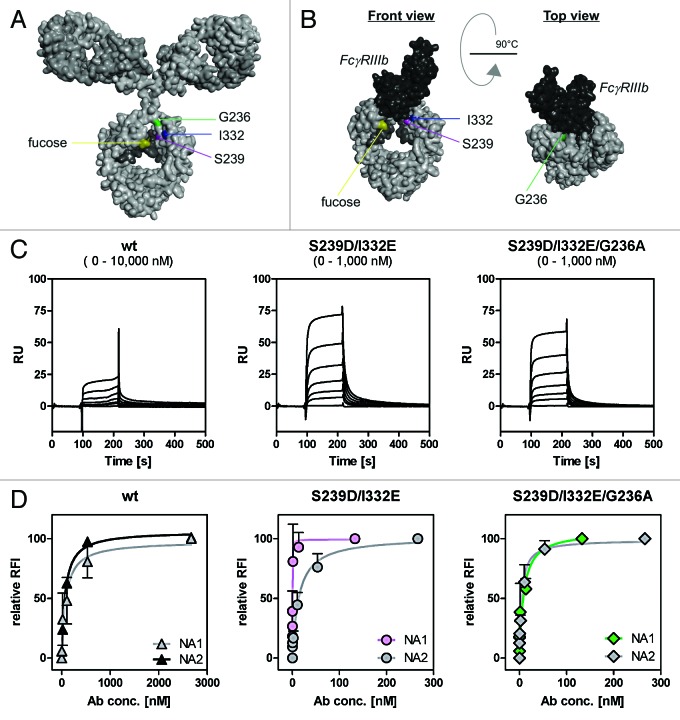
Figure 4. Determination of binding capacities of distinct antibody variants to FcγRIIIb. (A) Structural model of human IgG1 (pdb file from Clark, MR, Chem Immunol 1997; 65:88–110). The presented model illustrates the position of analyzed amino acid substitutions, which are located in the CH2 domain, as well as the fucose residues in IgG1. (B) Crystal structure of FcγRIIIb in complex with an Fc fragment of IgG1 (pdb file IT89). Pictures in A and B were generated using Discovery Studio 3.5 Client software (Accelrys). (C) SPR analyses were performed to determine binding capacities of distinct analyzed antibody variants to FcγRIIIb. One out of three independent experiments is presented per antibody variant. (D) Dose-response curves were prepared for analyzed antibody variants at indicated concentrations using CHO cell lines, either stably transfected with plasmids encoding for FcγRIIIb-NA1 or FcγRIIIb-NA2. Means ± SEM of at least three independent experiments are presented.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.