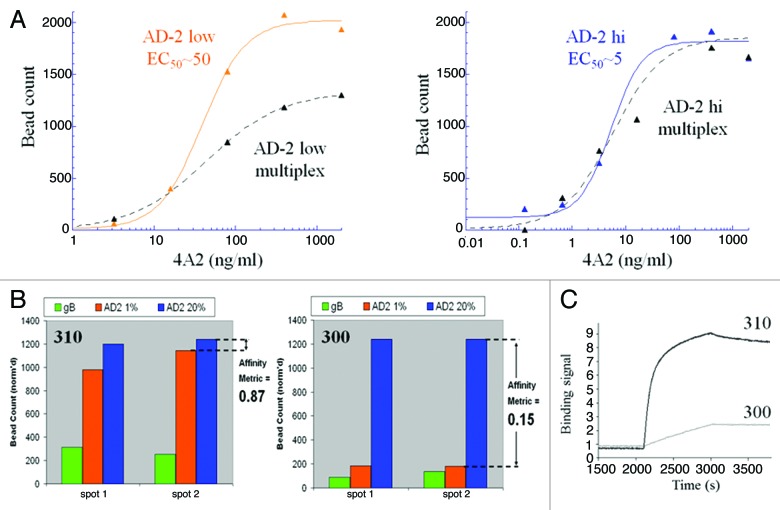
Figure 4. Development of a multiplexed strategy for discovery of high-affinity antibodies to the neutralizing site I, AD-2 region of the gB protein on human CMV. (A) A bead-based ELISA was used to track the optimization of low and high density AD-2 beads with a 10-fold binding difference to a benchmark reagent, mAb 4A2. As desired, binding of 4A2 to the low density beads was reduced when multiplexed with high density and gB beads, whereas high density beads bound 4A2 equally in single or multiplexed formats. With these bead properties we expected to be able to screen and differentiate mAb at least 10-fold improved in AD-2 binding over 4A2. (B) Digital fluorescent microscopy was used to count and quantify antibody binding to low affinity beads (high density, 20% AD-2 coating in 1% BSA), high-affinity beads (low density, 1% AD-2 coating in 1% BSA) and gB beads. Data was transformed to a bar graph format and an “affinity metric” value was assigned. A high-affinity antibody would bind equally, without bias, to both densities of AD-2 coated beads and have a calculated “affinity metric” of 1. Low-affinity antibodies would preferentially bind the high density AD-2 coated beads, and demonstrate affinity metric values < 1. Antibodies that cannot bind the peptide in the context of the native gB protein will show no signal on the gB-ECD coated beads. (C) Kinetic AD-2 binding sensorgrams for the high- (affinity metric = 0.87) and low- (affinity metric = 0.15) affinity monoclonal antibodies corresponding to these CellSpot™ phenotypes.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.