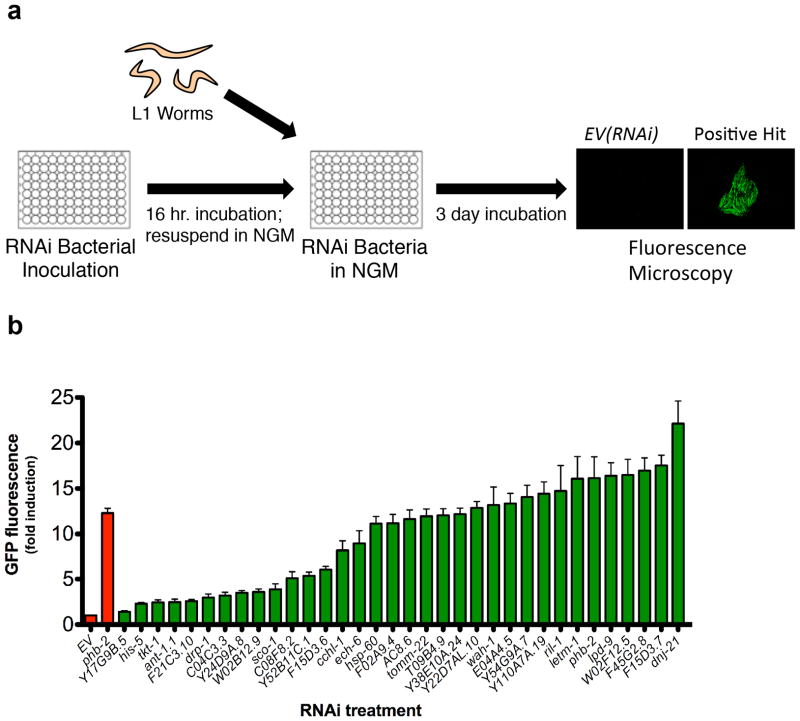
Figure 1. A genome-wide RNAi screen for negative regulators of the mitochondrial unfolded protein response.
(a) RNAi bacteria were grown overnight in 96 well plates, while hsp-6p::gfp animals were synchronized at L1 larval stage. The next day, RNAi bacteria was induced with IPTG, resuspended in liquid NGM, and added to reporter animals in 96-well plates. Animals were allowed to develop for three days and GFP was measured by fluorescent microscopy. (b) hsp-6p::gfp induction was quantified for 34 RNAi clones corresponding to positive hits that were not annotated as functioning in the ETC or mitochondrial translation. Validation included sequencing each RNAi clone and GFP quantification of individual animals grown at 20°C. GFP fluorescence is the mean fluorescence relative to EV(RNAi) (N=3 independent experiments, error bars indicate SEM).