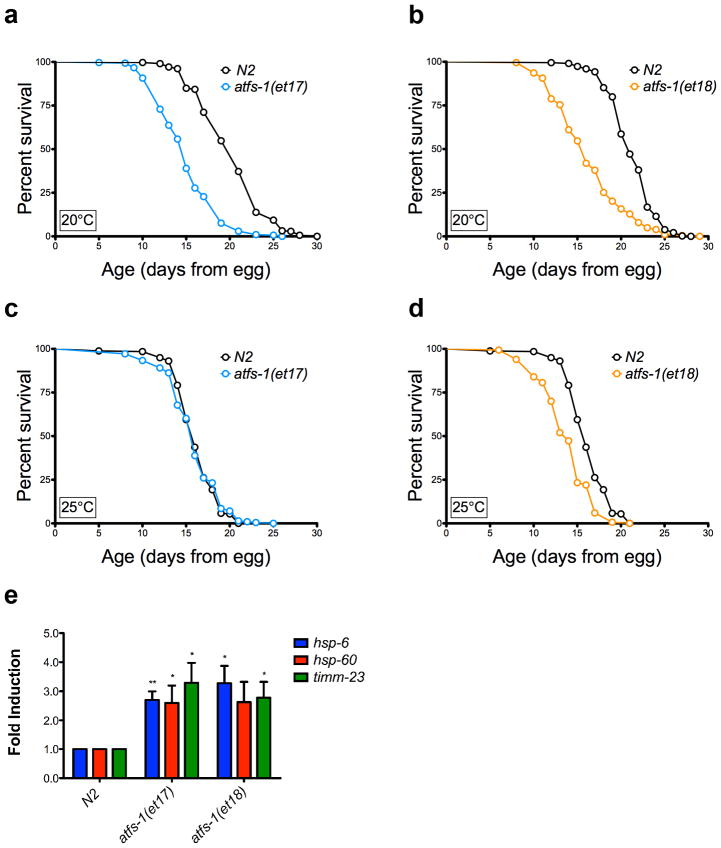
Figure 7. The UPRmt is not sufficient for lifespan extension.
(a) Constitutive UPRmt mutation atfs-1(et17) shortens lifespan at 20°C. N2 fed HT115 bacteria (mean 20.3 ± .2 days, n = 312), atfs-1(et17) fed HT115 bacteria (mean 15 ± .2, n = 303, p < 0.0001). (b) Constitutive UPRmt mutation atfs-1(et18) shortens lifespan at 20°C. N2 fed HT115 bacteria (mean 21.3 ± .1, n = 418), atfs-1(et18) fed HT115 bacteria (mean 16.3 ± .3 days, n = 203, p < 0.0001). (c) atfs-1(et17) mutation does not affect lifespan at 25°C. N2 fed HT115 bacteria (mean = 16.2 ± .15 days), atfs-1(et17) fed HT115 bacteria (mean = 15.9 ± .21 days, p = 0.31). (d) atfs-1(et18) mutation shortens lifespan at 25°C. N2 fed HT115 bacteria (mean = 16.2 ± .15 days), atfs-1(et18) fed HT115 bacteria (mean = 13.8 ± .13 days, p < 0.0001). (e) Constitutive UPRmt mutations atfs-1(et17) and atfs-1(et18) induce expression of hsp-6, hsp-60, and timm-23. Gene expression was normalized to N2 fed EV(RNAi). (N=4, Error bars represent SEM, p* < 0.05, p** < 0.01, student’s t-test). The gene expression data for hsp-60 in the atfs-1(et18) allele is trending significant, with p = 0.05. Lifespan experiments in this figure represent pooled data, are indicated as mean +/− SEM, and p-values were calculated using Wilcoxon rank-sum test. Data by individual experiment and statistical analysis provided in Supplementary Materials (Supplementary Data 2).