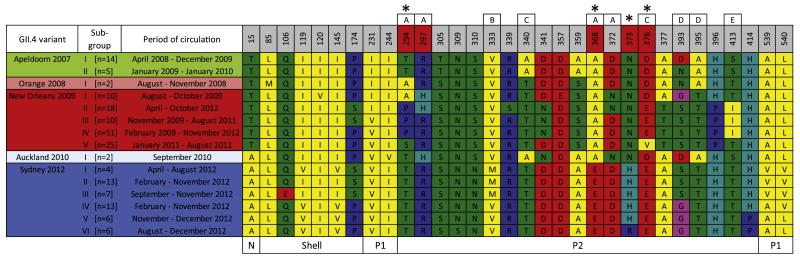
Fig. 4. Antigenic variation in the GII.4 Apeldoorn-lineage.
The GII.4 capsid sequence alignment of variants in the Apeldoorn-lineage used for the temporal evolutionary analysis was examined for antigenic clusters within and between different GII.4 variants as well as for evidence of positive selection. Asterisks indicate sites – 294, 368, 373 and 376 – that are under significant positive selection. Labelled boxes above each position indicate sites within the known blockade epitopes A–E that are important determinants of viral antigenicity. Amino acids have been coloured based on the properties of their side-chains: blue for positive charged – R, H, K; red for negative charged – D, E; Green for polar uncharged – S, T, N, Q; yellow for hydrophobic – A, V, I, L, M, F, Y, W; pink for special cases – C, U, G, P.