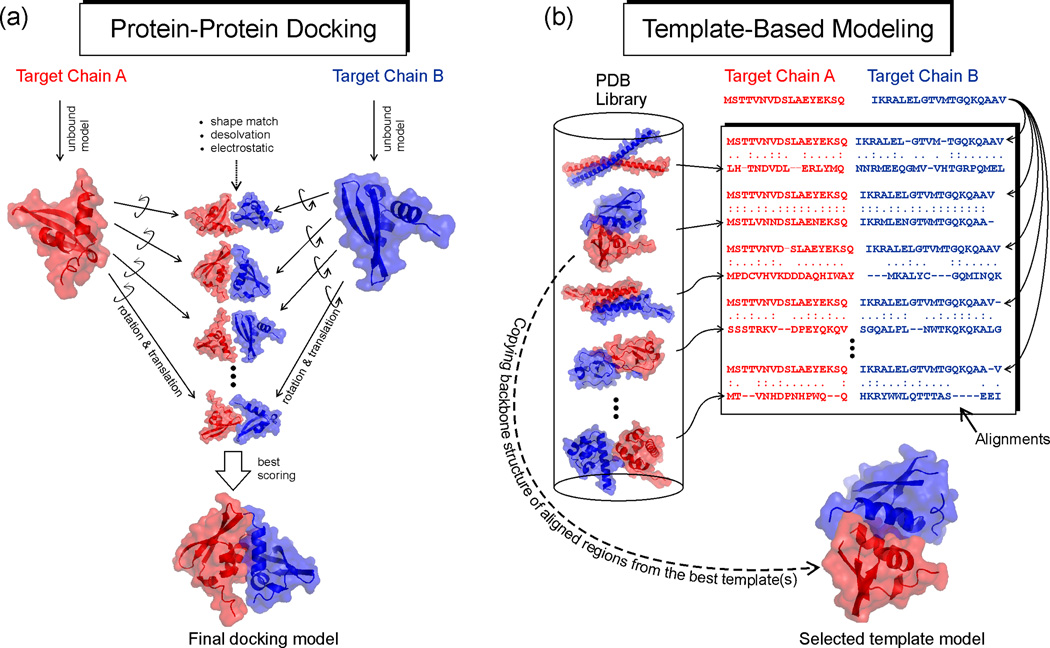
Figure 1.
Two principal protocols for protein complex structure prediction. Red and blue represent sequences and structures of two individual chains. (a) Rigid-body protein-protein docking constructs protein complex structures by assembling known structures of monomer components which are usually solved (or modeled) in their unbound states. The final model is selected from those with the best shape complementarity, desolvation free energy and electrostatic matches between interfaces of the component structures [9–12]. (b) Template-based modeling (TBM) identifies complex structure templates by aligning the amino acid sequences of the target chains with the solved complex structures in the PDB library (shown on the left). The alignment can be generated based on sequence, sequence profile, or a combination of the sequence and structure feature information. The best template of the highest alignment score is selected; and the structure framework in the aligned regions is copied from the template protein which serves as a basis for constructing the structure model of the target [18•,21••,24–25]. Note that (b) only shows a typical protocol of homology-based template detection. There are variants of TBM which detect complex templates by query and template structure comparisons (see Figure 2) [19•–20••,22•–23,30].