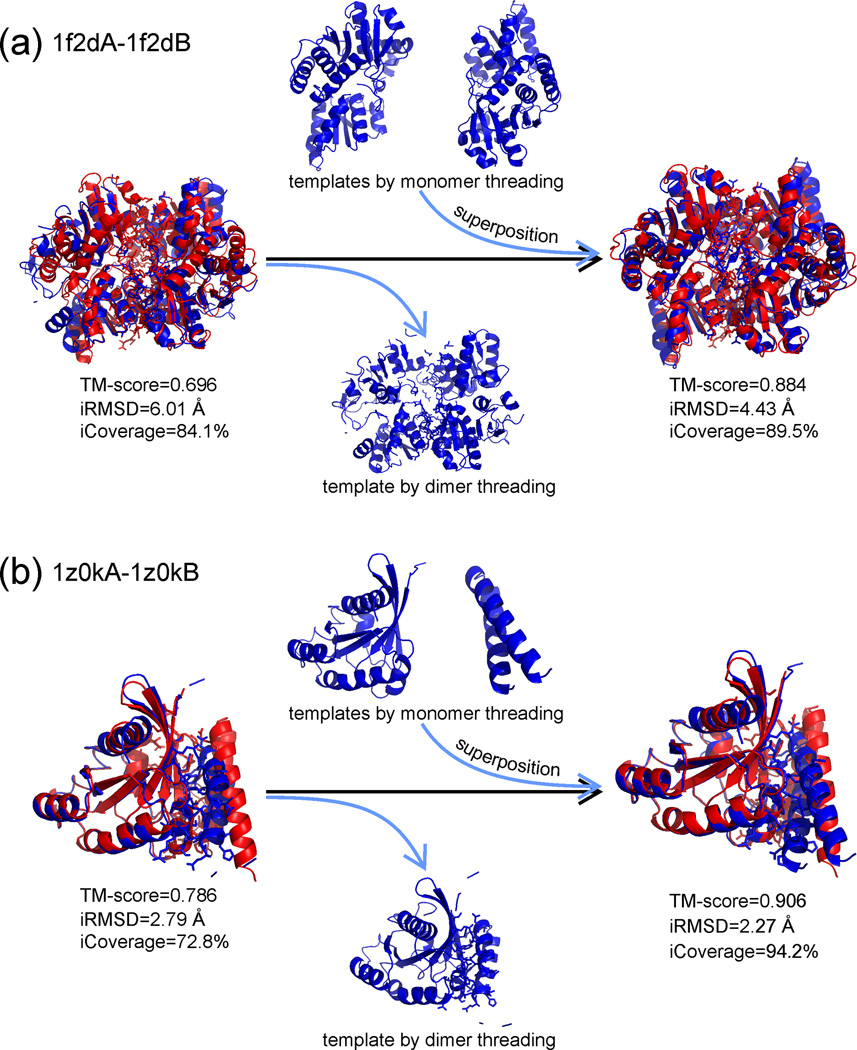
Figure 3.
Tertiary structure models from monomer threading were used to improve the model accuracy of dimeric threading models by structural superposition in COTH [18•]. Red cartoons represent experimental structures and blue ones are predicted models from monomer and dimeric threading, with sticks highlighting the interface residues. (a) A homodimer example from the 1-aminocyclopropane-1-carboxylate deaminase (PDB ID: 1f2d), which has the TM-score increased from 0.696 to 0.884 after the structural superposition of the monomer threading models on the dimer threading framework. The interface RMSD (iRMSD) is reduced from 6.01 Å to 4.43 Å with the alignment coverage of interface residues (iCoverage) increasing from 84.1% to 89.5%. (b) A heterodimer example from GTP-Bound Rab4Q67L GTPase (PDB ID: 1zok), where TM-score, iRMSD and iCoverage are improved, after the structure superposition, from 0.786, 2.79 Å, 72.8% to 0.906, 2.27 Å and 94.2%, respectively.