SUMMARY
MALT1 cleavage activity is linked to the pathogenesis of activated B cell-like diffuse large B cell lymphoma (ABC-DLBCL), a chemoresistant form of DLBCL. We developed a MALT1 activity assay and identified chemically diverse MALT1 inhibitors. A selected lead compound, MI-2, featured direct binding to MALT1 and suppression of its protease function. MI-2 concentrated within human ABC-DLBCL cells and irreversibly inhibited cleavage of MALT1 substrates. This was accompanied by NF-κB reporter activity suppression, c-REL nuclear localization inhibition, and NF-κB target gene downregulation. Most notably, MI-2 was nontoxic to mice, and displayed selective activity against ABC-DLBCL cell lines in vitro and xenotransplanted ABC-DLBCL tumors in vivo. The compound was also effective against primary human non-germinal center B cell-like DLBCLs ex vivo.
INTRODUCTION
Non-Hodgkin’s lymphoma (NHL) is the seventh most frequent cancer (Siegel et al., 2012). Diffuse large B cell lymphoma (DLBCL) is the most common subtype of NHL, accounting for ~25% of all lymphoma cases (Swerdlow et al., 2008). Gene expression profiling allowed subclassification of DLBCL into distinct molecular subtypes, including germinal center B cell-like (GCB) DLBCL, activated B cell-like (ABC) DLBCL, and primary mediastinal B cell lymphoma (PMBL) (Alizadeh et al., 2000; Rosenwald et al., 2003). These subtypes differ significantly in their spectrum of recurrent somatic mutations, dependence on different signaling pathways, and response to current standard therapies (Lenz et al., 2008b; Wright et al., 2003). Patients with the GCB subtype have a significantly better overall survival compared to those with the ABC subtype (Alizadeh et al., 2000; Rosenwald et al., 2002). Improved therapies are needed for all DLBCLs but most urgently for ABC-DLBCLs, which are the most chemoresistant.
ABC-DLBCL is characterized by its reliance on the oncogenic activation of the NF-κB pathway through several different mechanisms. These mostly involve somatic mutations in molecules participating in signaling downstream of the B cell receptor (BCR), including activating mutations of CARMA1/CARD11 (Lenz et al., 2008a) and CD79A/B (Davis et al., 2010), homozygous deletion/inactivating mutations of TNFAIP3/A20 (Compagno et al., 2009; Honma et al., 2009), and activating mutations of MYD88 downstream of the Toll-like receptor (Ngo et al., 2011). CARMA1 forms part of the CARMA1-BCL10-MALT1 (CBM) complex and mediates NF-κB activation downstream of the B cell receptor, T cell receptor (Ruefli-Brasse et al., 2003; Ruland et al., 2003), and ITAM-coupled natural killer cell receptors (Gross et al., 2008). The MALT1 (mucosa-associated lymphoid tissue lymphoma translocation 1) subunit is the active signaling component of the CBM complex (Lucas et al., 2001) and features protease activity that cleaves and inactivates inhibitors of the NF-κB signaling pathway such as TNFAIP3/A20 (Coornaert et al., 2008), CYLD (Staal et al., 2011), and RELB (Hailfinger et al., 2011) or the BCL10 protein (Rebeaud et al., 2008), indirectly activating NF-κB signaling. MALT1 translocations, including t(11;18)(q21;q21), which produces an API2-MALT1 fusion, and t(14;18)(q32;q21), which results in the IGH-MALT1 translocation, are detected in up to 55% of MALT-type lymphomas (Farinha and Gascoyne, 2005). These translocations lead to overexpression of MALT1 and, in the case of the API2-MALT1 translocation, constitutive activation of the pathway (Dierlamm et al., 1999; Sanchez-Izquierdo et al., 2003; Streubel et al., 2003). Constitutive expression of MALT1 in mice induces a disease that is similar to MALT lymphomas in humans, and induces ABC-like DLBCLs in a p53-null background (Vicente-Dueñas et al., 2012). MALT1 has not been found mutated or translocated in DLBCL but is gained along with BCL2, and this low-copy-number amplification is associated with an ABC-DLBCL phenotype (Dierlamm et al., 2008). Moreover, ABC-DLBCL cell lines have been shown to be dependent on MALT1 catalytic activity (Ferch et al., 2009; Hailfinger et al., 2009; Ngo et al., 2006).
MALT1 is a paracaspase, which is related to the caspase (cysteine-aspartic proteases) family of proteases but cleaves after Arg residues instead of Asp (Rebeaud et al., 2008). MALT1 is the only gene encoding paracaspase in the human genome. MALT1-null animals display defects in B and T cell function but are otherwise healthy (Ruefli-Brasse et al., 2003; Ruland et al., 2003). These factors suggest that MALT1-targeted therapy would likely be well tolerated with little or manageable toxicity. Consequently, MALT1 represents a potentially important therapeutic target for ABC-DLBCL and MALT lymphoma.
RESULTS
Biochemical Screening Identifies Low Molecular Weight Inhibitors of MALT1 Proteolytic Activity
We reasoned that MALT1 small molecule inhibitors might be useful chemical tools for studying MALT1 biology and treating MALT1-addicted tumors. However, full length MALT1 and its paracaspase domain (amino acids 340–789) are naturally present in physiological solutions as a monomer, which has very low proteolytic activity. Caspases generally must homodimerize for maximal catalytic activity (Pop et al., 2006; Walker et al., 1994; Yin et al., 2006), and accordingly the recently reported structures of the paracaspase domain of MALT1 in complex with a peptide inhibitor are dimeric (Wiesmann et al., 2012; Yu et al., 2011). In order to generate catalytically active MALT1 for an effective assay to screen for inhibitors, we biochemically engineered a recombinant form of MALT1 (340–789) fused with a leucine zipper dimerization motif (LZ-MALT1), which promotes its dimerization and activation (Figure 1A). We developed a MALT1 activity assay using the MALT1 substrate peptide LRSR linked to the fluorogen AMC (7-amino-4-methylcoumarin). Cleavage of the Ac-LRSR-AMC substrate by MALT1 resulted in release of AMC and a fluorescent signal.
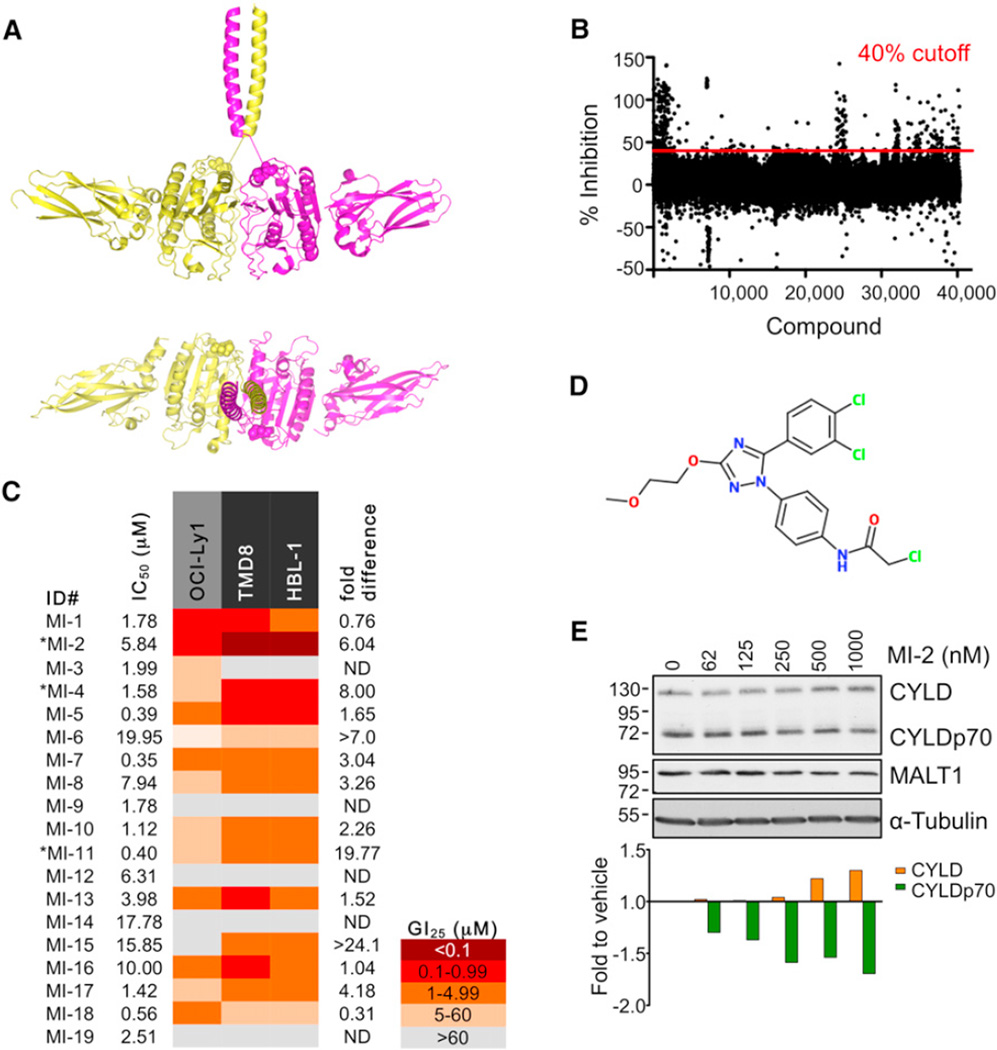
Figure 1. Identification of MALT1 Small Molecule Inhibitors.
(A) Dimeric LZ-MALT1 representation showing LZ-MALT1 monomers in yellow and magenta. Met338 is shown as a sphere model. The model was generated using the MALT1 structure (Protein Data Bank [PDB] ID code 3UOA) and LZ structure (PDB ID code 2ZTA).
(B) Represented is the % inhibition for each of the compounds assayed at 12.5 µM. Cutoff was 40% inhibition.
(C) Summary table of IC50 and GI25 results for screening hits. Experiments were performed three times in triplicate. Fold difference = OCI-Ly1 GI25/average GI25 for the MALT1-dependent cell lines. *Significant dose-dependent suppression of proliferation in ABC-DLBCL, regression extra sum-of-squares F test.
(D) MI-2 structure.
(E) MALT1 cleavage of CYLD inhibition by MI-2 in HBL-1 cells at 24 hr studied by western blot. α-tubulin, and loading control. Densitometry values were normalized to α-tubulin and are relative to vehicle-treated cells. The representative result is from three experiments.
See also Figure S1.
The optimal conditions for high-throughput screening were determined by systematic variation of the concentrations of the enzyme and the substrate in a two-dimensional grid. Fluorescence measurements were taken every 45 s for 60 min. The measurements were plotted as a function of time. Conditions with a linear relationship between fluorescence and time were considered appropriate for screening. Quality was assessed using the Z′ factor, a coefficient reflective of the dynamic range of the assay and variance of the data (Zhang et al., 1999), calculated by the formula Z′ factor = 1 − 3 × (σp + σn)/(|μp − μn|), where σp/n is the standard deviation for positive and negative control and μp/n is the mean for positive and negative control. The Z′ factor for this screen was 0.738, which is within the optimal range 0.5–1 (Zhang et al., 1999). A total of 46,464 compounds was screened (see **Supplemental Experimental Procedures available online). Using 40% inhibition as a threshold, 324 candidate compounds were selected for validation in a concentration-response assay (Figure 1B). Of these, 19 compounds (Figure S1A) were selected for further validation based on their biochemical activity (half-maximal inhibitory concentration [IC50] < 20 µM; Figure 1C).
Candidate Inhibitors Selectively Suppress ABC-DLBCL Cell Lines and MALT1 Catalytic Activity
MALT1 activity plays an important role in selectively maintaining proliferation of ABC-DLBCL cell lines (Ngo et al., 2006). Accordingly, ABC- and GCB-DLBCL cell lines present differential sensitivity to MALT1 cleavage inhibition by the peptide Z-VRPR-FMK (Ferch et al., 2009; Hailfinger et al., 2009; Rebeaud et al., 2008). To determine whether candidate small molecules display a similar profile, two ABC-DLBCL cell lines, HBL-1 and TMD8, and one GCB-DLBCL cell line, OCI-Ly1, were exposed to increasing concentrations of the 19 selected molecules. Cell proliferation was measured 48 hr after exposure to a single dose of compound using an ATP-based metabolic luminescent assay (summarized in Figure 1C). Three compounds consistently induced significant selective dose-dependent suppression of ABC-DLBCL cells (MI-2, p < 0.0001; MI-4, p = 0.006; MI-11, p < 0.0001; regression extra sum-of-squares F test; Figures 1C and S1B). Hence, these compounds were active in cells, selective for ABC-DLBCLs, and lack nonspecific cellular toxicity. MI-6 and MI-15 also showed differential inhibition of ABC-DLBCL cells but did not reach statistical significance.
Compound MI-2 was the most potent in cell-based assays, with 25% growth-inhibitory concentration (GI25) values in the high-nanomolar range. MI-2 (Figure 1D) was therefore next assayed for inhibition of MALT1-mediated substrate cleavage in lymphoma cells. HBL-1 cells were treated with increasing concentrations of MI-2 for 24 hr and cleavage of the MALT1 target protein CYLD was measured by western blotting and densitometry. MI-2 caused a dose-dependent decrease in MALT1-mediated cleavage, noted by an increase in the uncleaved CYLD protein and a decrease in the cleaved form of the protein (Figure 1E). MI-2 was selective as a MALT1 paracaspase inhibitor, because it displayed little activity against the structurally related caspase family members caspase-3, -8, and -9 (Figure S1C). Moreover, MI-2 did not inhibit caspase-3/7 activity or apoptosis in cell-based assays at concentrations that suppress MALT1 (Figures S1D–S1F). Hence, MI-2 is a potential lead compound as a therapeutic MALT1 inhibitor.
MI-2 Analogs Display MALT1-Inhibitory Activity
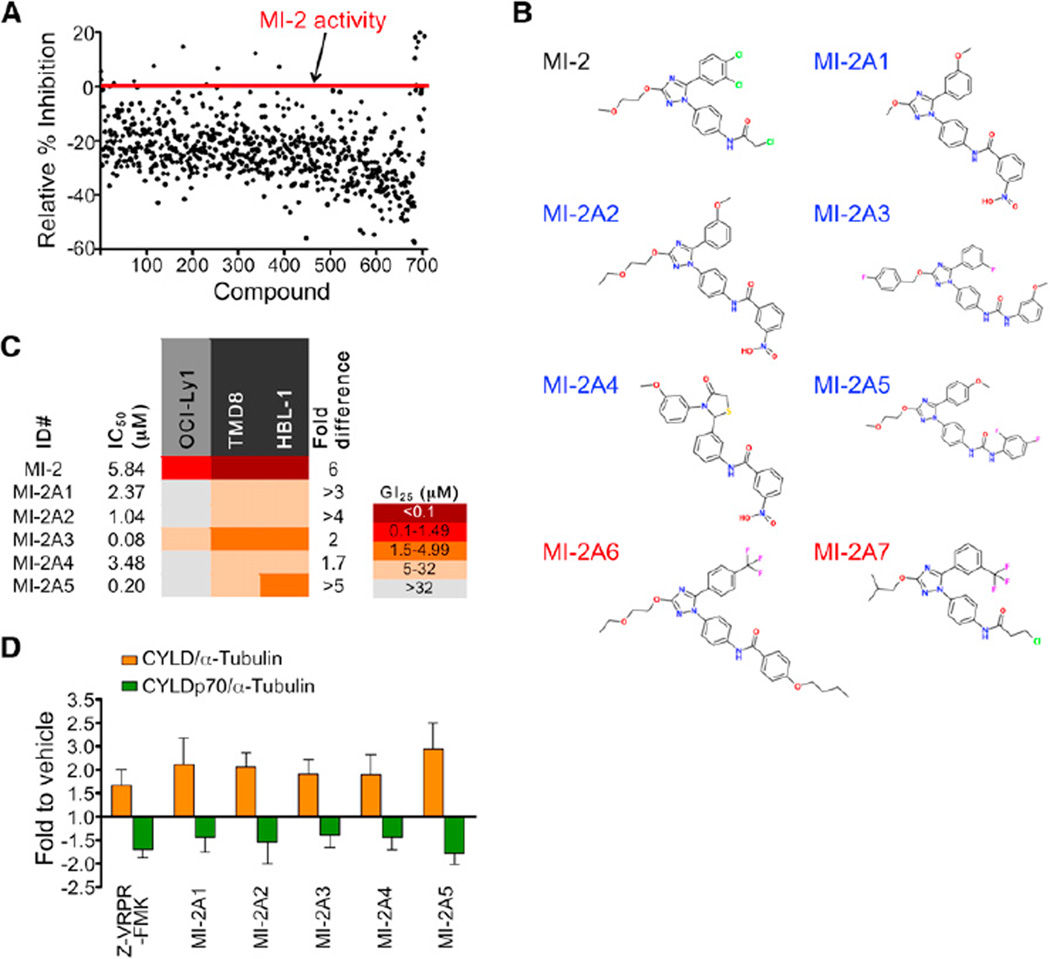
To establish whether compound MI-2 represented a potential scaffold for development of MALT1 inhibitors, we compared MI-2 with other chemical compounds in silico to identify potential analogs. A total of 704 analog compounds from available libraries with similarity score ≥70% (Tanimoto similarity functions) was screened by LZ-MALT1 fluorescence assay. Nineteen analogs displaying equal or higher activity than MI-2 were selected (Figure 2A). Five analogs with biochemical IC50s within a similar range as MI-2 were selected for further characterization in cell-proliferation assays (Figures 2B and 2C). All five analogs (MI-2A1–5) exhibited the same trend toward selective suppression of the ABC-DLBCL cell lines, with GI25 concentrations in the micromolar range (Figure 2C). Two analog compounds with no LZ-MALT1-inhibitory activity in vitro (MI-2A6 and -7) used as chemical controls had no effect on cell proliferation over the same dose range (Figure S2). The five active MI-2 analogs were assayed for inhibition of MALT1 cleavage of CYLD. All five compounds, administered at 5 µM for 8 hr, showed cleavage inhibition similar to the Z-VRPR-FMK MALT1 blocking peptide (50 µM) used as positive control (Figure 2D), although MI-2 itself remained the most potent compound. Collectively, the conservation of MALT1 inhibitor activity in vitro and in cell-based assays among chemically related compounds points toward the suitability of MI-2 and its analogs as lead compound inhibitors of MALT1.
Figure 2. MI-2 Analogs Display Similar MALT1-Inhibition Activity.
(A) Seven hundred and four compounds with over 70% homology to MI-2 were screened. Compounds with equal or higher activity than MI-2 were selected.
(B) Structures of the MI-2 analog compounds assayed in cell growth-inhibition experiments. Blue, active analogs; red, inactive analogs.
(C) IC50 and GI25 values for the selected analogs assayed in HBL-1, TMD8, and OCI-Ly1 cells. Experiments were performed three times in triplicate. Fold difference = OCI-Ly1 GI25/average GI25 for the MALT1-dependent cell lines.
(D) CYLD cleavage was studied in HBL-1 cells treated with 5 µM analog compound or 50 µM Z-VRPR-FMK for 8 hr. Densitometry results were normalized to α-tubulin and fold change-to-vehicle ratios were calculated. Results are mean ± SEM of three independent experiments.
See also Figure S2.
MI-2 Directly Binds and Irreversibly Inhibits MALT1
We next investigated whether MI-2 directly bound to MALT1 or indirectly affected MALT1 activity, for example through binding to the LZ region of the fusion protein. Heteronuclear single-quantum coherence (HSQC) nuclear magnetic resonance (NMR) spectroscopy was used to characterize the binding of MI-2 to the paracaspase domain of MALT1 (residues 329–728). As MI-2 was titrated in, resonances corresponding to the unbound state of MALT1 decreased in intensity, while another set of resonances corresponding to the MALT1-MI-2 complex gradually appeared (Figure 3A). This pattern of chemical shift changes is characteristic of slow exchange on the NMR timescale and is indicative of a robust interaction between MALT1 and MI-2. In contrast, NMR spectroscopy studies showed no evidence of binding by the inactive analogs MI-2A6 and MI-2A7 (Figure 3B).
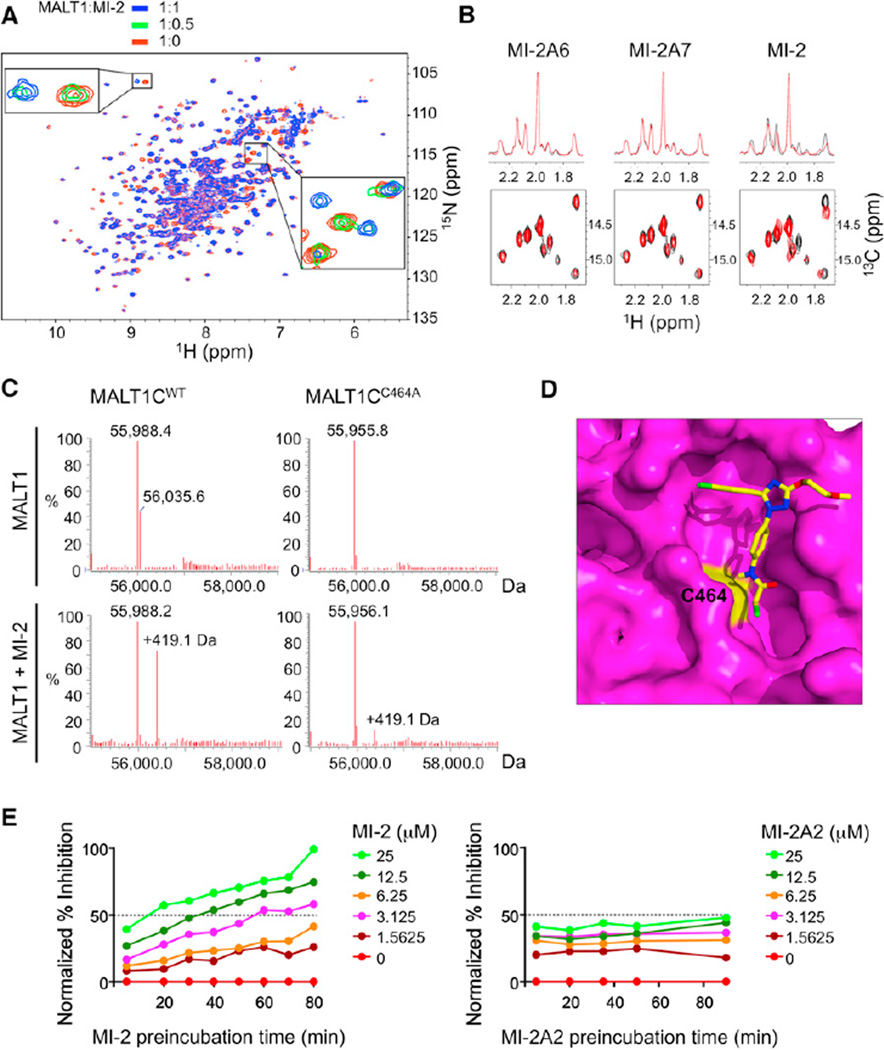
Figure 3. MI-2 Directly Interacts with and Irreversibly Inhibits MALT1.
(A) Superposition of the 1H-15N HSQC spectrum of the apo-MALT1 (red) with the MALT1-MI-2 complex (blue) at a molar ratio of 1:1. The expanded regions highlight interacting residues in the slow-exchange regime on the NMR timescale (intermediate 1:0.5 MALT1:MI-2 ratio is shown in green).
(B) One-dimensional (top) and two-dimensional (bottom) 1H-13C HSQC NMR spectra of MALT1 (329–728) without (black) and with compounds MI-2A6, MI-2A7, and MI-2 (red).
(C) LC-MS for MALT1WT and MALT1C464A (amino acids 329–728) with and without MI-2.
(D) Docked MI-2 (in stick model) on the MALT1 paracaspase domain (in surface representation). MALT1 is shown in magenta with C464 in yellow. MI-2 is shown with carbons in yellow, oxygens in red, nitrogens in blue, and chlorines in green.
(E) LZ-MALT1 was preincubated with the indicated concentrations of MI-2 or MI-2A2 (0–25 µM) for different durations (5–90 min) before Ac-LRSR-AMC was added. The graphs represent normalized% inhibition compared to preincubation time.
See also Figure S3.
Because MI-2 contains a reactive chloromethyl amide, we investigated whether MI-2 could modify MALT1 covalently using liquid chromatography-mass spectrometry (LC-MS). As shown in Figure 3C, MALT1 paracaspase domain (329–728) presented a major peak at 55,988.4 Da. Upon incubation with the compound MI-2, the major peak of MALT1 was shifted to 56,407.5 Da, an increase of 419.1 Da. This corresponds to addition of MI-2 minus the chloride group, indicating that MI-2 can bind covalently to MALT1 and potentially act as an irreversible inhibitor. Because the chloromethyl amide group is not conserved in the active MI-2 analogs (Figure 2B), it is most likely the common chemical scaffold in the MI-2 series that provides specificity to MALT1. Notably, LC-MS performed with MI-2 and the MALT1 active site mutant C464A revealed markedly reduced covalent binding, suggesting that the active site C464 residue is the main target of modification by MI-2 (Figure 3C). To further explore the potential mode of binding of MI-2 to the MALT1 paracaspase domain, we employed molecular docking using AutoDock 4.2 (Morris et al., 2009). The crystal structure of MALT1 (Wiesmann et al., 2012; Yu et al., 2011) was kept as a rigid body while allowing conformational flexibility of MI-2. The final results were ranked on the predicted binding free energy and the cluster size for each docking conformation. The top five poses were selected, all of which had similar docking scores with slight changes in their orientations. As shown for the first top hit, MI-2 appears to bind the active site cleft with its chloromethyl group close to the active site C464 in the paracaspase domain (Figures 3D and S3A), consistent with a covalent-bond formation between these two groups. Collectively, the data suggest that MI-2 engages and irreversibly binds the MALT1 active site.
To examine whether MI-2 inhibition of MALT1 is consistent with irreversible binding kinetics, LZ-MALT1 was preincubated with different concentrations of MI-2 for 5–80 min followed by addition of the substrate Ac-LRSR-AMC to determine enzymatic activity (Figures 3E and S3B). Notably, the percent MALT1 inactivation increased with time, reaching plateaus near the end of the test, consistent with covalent and irreversible inhibition. Inhibition was concentration dependent, with higher concentrations showing greater inactivation and faster rates of saturation. In contrast, the active MI-2 analog MI-2A2, which does not have the chloromethyl amide group, showed no evidence of cumulative inhibition of MALT1, consistent with reversible inhibition. It should be noted that MI-2 reached close to 100% inhibition, whereas MI-2A2 with a lower IC50 only reached ~50% inhibition (Figure 3E). The irreversible kinetics might contribute to the more potent effects of MI-2 in cell-based assays versus its analogs that lack the chloromethyl amide group and only bind reversibly, as has been noted in the case of peptidyl halomethyl ketone protease inhibitors (Powers et al., 2002).
MI-2 Inhibits MALT1 Functions in ABC-DLBCL Cell Lines
Having confirmed MI-2 as a lead compound, we next explored its effects on MALT1 signaling in ABC-DLBCL cells. We first examined the impact of MI-2 on cleavage of additional MALT1 substrates such as A20, BCL10, and RELB. Because these proteins are directed to proteasomal degradation after cleavage (Coornaert et al., 2008; Hailfinger et al., 2011; Rebeaud et al., 2008), we used the proteasome inhibitor MG-132 to facilitate visualization of cleavage products (Figure 4A). HBL-1 and TMD8 cell lines were exposed to either MI-2 (2 µM) or vehicle for 30 min followed by 5 µM MG-132 for an additional 1 (lanes 2 and 3) or 2 hr (lanes 4 and 5) in order to allow cleaved forms of MALT1 substrates to accumulate during exposure to MI-2. As expected, MG-132 exposure revealed the accumulation of A20, BCL10, and RELB cleavage products due to the constitutive activity of MALT1 in these DLBCL cells. However, exposure to MI-2 diminished the abundance of cleaved forms and/or increased the abundance of full-length proteins, consistent with the loss of MALT1 enzymatic activity (Figure 4A).
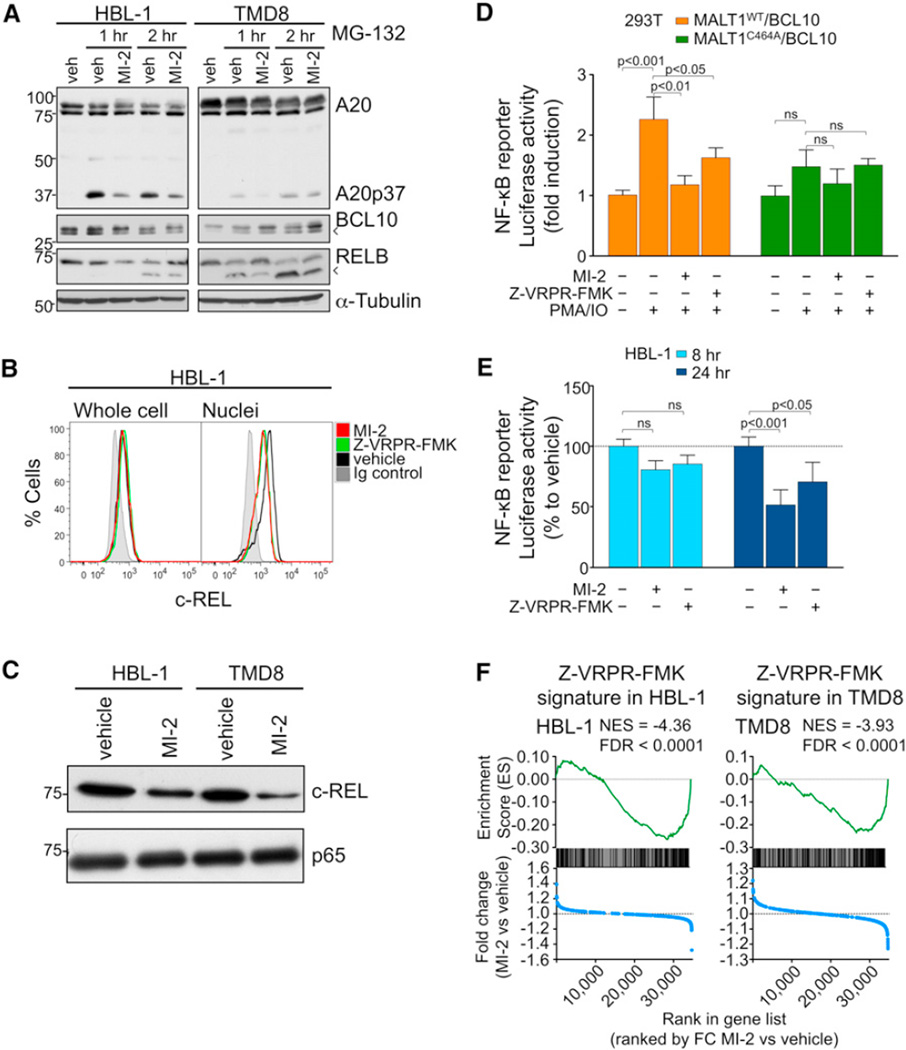
Figure 4. MI-2 Inhibits MALT1 Signaling in DLBCL Cells.
(A) Western blots for A20, RELB, and BCL10 after 30 min pretreatment with 2 µM MI-2 or vehicle, followed by proteasome inhibitor MG-132 (5 µM) treatment for 1 or 2 hr in HBL-1 and TMD8. Arrowheads indicate cleavage products.
(B) Flow cytometry for c-REL whole cells or isolated nuclei from HBL-1 cells treated with 200 nM MI-2, 50 µM Z-VRPR-FMK, or vehicle for 24 hr.
(C) c-REL and p65 protein expression in nuclear extracts of HBL-1 and TMD8 cells treated for 24 hr with 200 or 500 nM MI-2, respectively.
(D) NF-κB reporter assays in 293T cells transfected with the indicated expression constructs. Cells were treated for 30 min with 2 µM MI-2, 50 µM Z-VRPR-FMK, or vehicle followed by PMA/ionomycin for 2 hr. The y axis represents luciferase fold induction relative to vehicle-treated cells, normalized to internal control. Statistics, ANOVA, Bonferroni posttest.
(E) NF-κB reporter assays in HBL-1 cells treated with 200 nM MI-2, 50 µM Z-VRPR-FMK, or vehicle for 8 and 24 hr. The y axis represents % luciferase activity relative to vehicle-treated cells, normalized to internal control. Statistics, ANOVA, Bonferroni posttest.
(F) Enrichment of Z-VRPR-FMK-downregulated genes after MI-2 treatment in HBL-1 and TMD8 cells using GSEA. Normalized enrichment score (NES) and false discovery rate (FDR) values are indicated. Top: enrichment score versus relative position of ranked genes (green tracing); bottom: fold change (FC) MI-2/vehicle versus relative position of ranked genes (blue tracing). Experiments were performed in triplicate unless otherwise specified. Data are mean ± SEM.
See also Figure S4.
MALT1 mediates c-REL translocation to the nucleus following BCR stimulation (Ferch et al., 2007). Therefore, HBL-1 cells were exposed to 200 nM MI-2, 50 µM Z-VRPR-FMK (positive control), or vehicle for 24 hr, followed by c-REL flow cytometry of whole cells or isolated nuclei. Both MI-2 and Z-VRPR-FMK reduced nuclear c-REL to a similar extent, without affecting whole-cell levels of this protein (Figure 4B). To further confirm this result, we also performed western blots for c-REL and p65 in nuclear extracts of HBL-1 and TMD8 cells treated for 24 hr with GI50 concentrations of MI-2 (200 nM for HBL-1 and 500 nM for TMD8). In both cell lines, exposure to MI-2 caused a clear reduction of nuclear c-REL whereas it did not affect p65 levels (Figure 4C). This selectivity toward c-REL had also been previously shown in MALT1 knockout mice and after MALT1 cleavage inhibition by the MALT1 blocking peptide Z-VRPR-FMK (Ferch et al., 2007, 2009; Hailfinger et al., 2011).
Antigen receptor-mediated NF-κB signaling partly depends on MALT1 activity (Ruefli-Brasse et al., 2003; Ruland et al., 2003). Hence, we tested the effect of MI-2 on attenuating NF-κB activation induced by phorbol 12-myristate 13-acetate (PMA)/ionomycin, which mimics BCR activation and activates MALT1-dependent cleavage (Coornaert et al., 2008; Rebeaud et al., 2008). First, 293T cells were transfected with the NF-κB reporter vector (NF-κB)5-luc2CP-pGL4 (harboring five copies of the NF-κB consensus-response element and a destabilized firefly luciferase) and TK-pRL control together with plasmids expressing BCL10 and either MALT1WT or MALT1C464A (inactive mutant). Exposure to PMA/ionomycin significantly increased luciferase activity in 293T cells when MALT1WT was transfected, but not with the mutant MALT1C464A. Pretreatment with MI-2 significantly inhibited NF-κB induction by PMA/ionomycin stimulation similarly to Z-VRPR-FMK, whereas it did not significantly affect that of MALT1C464A (Figure 4D). HBL-1 cells are reported to exhibit chronic active B cell receptor signaling with consequent NF-κB activation (Davis et al., 2010). HBL-1 was transfected with the reporter construct (NF-κB)5-luc2CP-pGL4 and TK-pRL control. Treatment with MI-2 promoted a 20% and 50% reduction in NF-κB reporter activity at 8 and 24 hr, respectively. A similar result was observed for Z-VRPR-FMK (50 µM) (Figure 4E). This reduction in NF-κB reporter activity was significant at 24 hr for MI-2 and the blocking peptide Z-VRPR-FMK.
The impact of MI-2 on NF-κB signaling was further characterized by gene expression profiling. For these experiments, the HBL-1 and TMD8 cell lines were treated with GI50 concentrations of MI-2 (HBL-1, 200 nM; TMD8, 500 nM) or 50 µM Z-VRPR-FMK for 8 hr, and RNA was extracted for gene expression studies using oligonucleotide microarrays. Z-VRPR-FMK was previously shown to attenuate the NF-κB signature in ABC-DLBCL cell lines (Hailfinger et al., 2009). MI-2 would be expected to exhibit a similar profile. For this study, we assigned Z-VRPR-FMK signatures by capturing the top 200 downregulated genes by Z-VRPR-FMK treatment compared to vehicle for each cell line. We next performed gene set enrichment analysis (GSEA) of this Z-VRPR-FMK signature against the differential expression of all genes preranked by fold change between MI-2- and vehicle-treated cells for each cell line. The Z-VRPR-FMK signature was significantly enriched among genes downregulated after MI-2 treatment for both cell lines (Figure 4F). GSEA was next performed using two independent ABC-DLBCL NF-κB gene expression signatures derived from either OCI-Ly3 and OCI-Ly10 or HBL-1 cell lines. We observed significant enrichment of these NF-κB gene sets among genes downregulated after MI-2 treatment in both cell lines (Figure S4). Collectively, these data suggest that MI-2 suppresses NF-κB activity induced by MALT1, similar to the effect observed with Z-VRPR-FMK.
MI-2 Selectively Suppresses MALT1-Dependent DLBCL Cell Lines
To further explore the spectrum of MI-2-mediated MALT1-inhibition effects, we turned to a larger panel of six ABC-DLBCL and two GCB-DLBCL cell lines (Table S1). Endogenous MALT1 activity was evaluated by western blotting for A20, BCL10, and CYLD, and NF-κB activation by phospho-IκB-α and total IκB-α (Figure S5A). Dependence on MALT1 proteolytic activity for proliferation was tested by 50 µM Z-VRPR-FMK treatment for 48 hr (Figure S5B). As expected, the two GCB-DLBCL cell lines (OCI-Ly7 and OCI-Ly1) did not display evidence of MALT1 or NF-κB signaling and did not respond to Z-VRPR-FMK. The U2932 and HLY1 ABC-DLBCL cell lines harbor mutations in TAK1 and A20, which activate NF-κB signaling downstream of MALT1. Hence, these two cell lines displayed relatively little response to Z-VRPR-FMK. In contrast, the ABC-DLBCL cells HBL-1, TMD8, OCI-Ly3, and OCI-Ly10 displayed evidence of MALT1 activity and inhibition of proliferation by Z-VRPR-FMK, indicating that these four cell lines are MALT1 dependent.
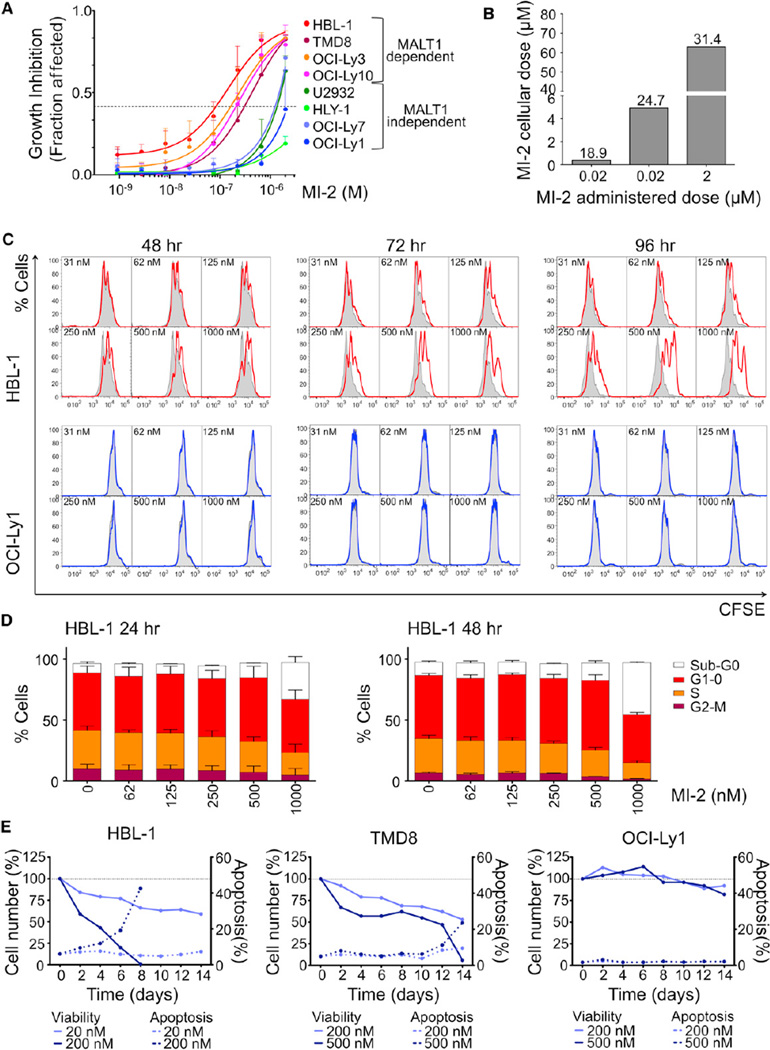
All eight cell lines were exposed to increasing concentrations of MI-2 (single dose) and cell proliferation was measured at 48 hr using an ATP-based metabolic luminescent assay (Figure 5A). Growth inhibition by MI-2 was selective for MALT1-dependent cell lines, whereas the ABC-DLBCL MALT1-independent cell lines, U2932 and HLY-1, and the two GCB-DLBCL cell lines were resistant. The GI50 for MI-2 in HBL-1, TMD8, OCI-Ly3, and OCI-Ly10 cells was 0.2, 0.5, 0.4, and 0.4 µM, respectively, which is lower than its IC50 in vitro (Figure 1). This is likely explained by the irreversible binding of MI-2 to MALT1 as shown in Figure 3, but could also be due to intracellular accumulation of the compound. Indeed, we observed an 18- to 30-fold increase in MI-2 intracellular concentration in experiments where HBL-1 cells were exposed to 0.02, 0.2, or 2 µM MI-2 for 2 hr and washed three times and MI-2 was measured by LC-MS (Figure 5B). The intracellular concentration in the 0.2 µM MI-2-treated cells was 5 µM, similar to the calculated in vitro IC50 (Figure 5B). To determine the kinetics of accumulation of free drug, we measured the intracellular concentration of MI-2 at the GI50 concentration of 0.2 µM at 30 min and 2, 6, 12, 24, and 48 hr (Figure S5C). By 12 hr, there was virtually no detectable free MI-2 within the cells. However, after exposure of HBL-1 cells to increasing concentrations of a single dose of MI-2, recovery of cells only started to become evident after 48 hr (of the 0.2 µM dose level; Figure S5D). These data suggest that the potent biological effects of MI-2 are due at least in part to its irreversible binding to MALT1 aided by its tendency to concentrate in cells.
Figure 5. MI-2 Inhibits Cell Proliferation of MALT1-Dependent DLBCL Cells.
(A) Dose-effect graphical representation for eight DLBCL cell lines exposed to MI-2 for 48 hr. Data are mean ± SEM of three independent experiments performed in triplicate.
(B) Intracellular MI-2 concentration determined by LC-MS in HBL-1 cells treated for 2 hr. The y axis represents MI-2 intracellular concentration. The x axis represents administered dose of MI-2. Fold-change accumulation in cells is indicated.
(C) CFSE dilution assay in HBL-1and OCI-Ly1 cells after treatment with MI-2 for 48, 72, and 96 hr. Cells (2 × 104) were measured; DAPI− cells were gated for analysis.
(D) BrdU pulse-labeled HBL-1 cells treated for 24 and 48 hr with MI-2 were analyzed by flow cytometry. The graph shows the % of cells for each cell-cycle phase for vehicle and each of the concentrations assayed. Data are mean ± SD of two independent experiments.
(E) Apoptosis time course in cell lines treated with MI-2 every 24 hr. Cells were counted using trypan blue and apoptosis was assessed by Annexin V+ DAPI− every 48 hr for a period of 14 days. Left y axis, % cell number relative to vehicle for each time point. Right y axis, % apoptotic cells for each time point.
See also Table S1 and Figure S5.
To explore in more detail the biological effects of MALT1 inhibition, HBL-1, TMD8, OCI-Ly10, and the GCB-DLBCL cell line OCI-Ly1 were treated with increasing concentrations of MI-2. Cell proliferation was examined using the 5-(and 6)-carboxyfluorescein diacetate succinimidyl ester (CFSE) dilution assay by flow cytometry on viable cells at 48, 72, and 96 hr. MI-2 substantially inhibited proliferation in HBL-1, TMD8, and OCI-Ly10 whereas it did not affect OCI-Ly1 (Figures 5C and S5E). Using BrdU incorporation-DAPI (4′, 6-diamidino-2-phenylindole) staining and flow cytometry to assess the cell cycle, it was evident that MI-2 induced a dose-dependent decrease in S phase, with a reciprocal increment in the proportion of cells in G1-0 and sub-G0 (Figure 5D). To determine whether MALT1 inhibitors induced apoptosis, the ABC-DLBCL cell lines HBL-1 and TMD8 were treated daily with MI-2 at their respective GI25 and GI50, and the control OCI-Ly1 cell line at the higher doses was used for TMD8. Trypan blue exclusion and apoptosis assessed by Annexin V+ DAPI− flow cytometry was measured every 48 hr for a period of 14 days. Whereas MI-2 had no effect on OCI-Ly1 cells, it profoundly suppressed both HBL-1 and TMD8 cells, with the former exhibiting earlier and higher abundance of apoptotic cells (Figure 5E). Using the more sensitive caspase-3/7 cleavage assay, we observed evidence of dose-dependent apoptosis within 48 hr in both ABC-DLBCL cell lines (Figure S5F). Hence, MI-2 powerfully suppresses the growth and survival of ABC-DLBCL cell lines.
Compound MI-2 Is Nontoxic to Animals
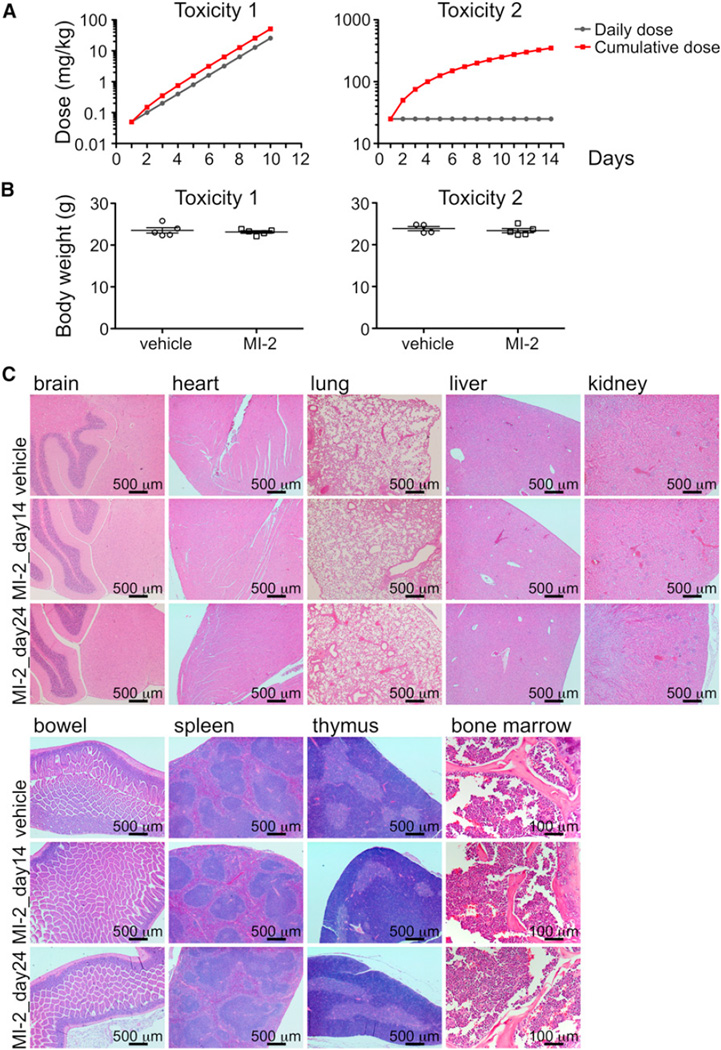
To determine its suitability as a MALT1 lead compound for in vivo studies, we examined whether MI-2 induced toxic effects in mice. Five C57BL/6 mice were exposed to daily intraperitoneal (IP) administration of increasing doses of MI-2 ranging from 0.05 to 25 mg/kg over the course of 10 days to a cumulative dose of 51.1 mg/kg, and another five mice were exposed to vehicle only (5% DMSO, n = 5) (Figure 6A, toxicity 1). There was no evidence of lethargy, weight loss (Figure 6B, toxicity 1), or other physical indicators of sickness. To ascertain whether the maximal administered dose of 25 mg/kg is safe in a 14 day schedule, we exposed ten mice to daily IP administration of 25 mg/kg of MI-2 over 14 days to a cumulative dose of 350 mg/kg, using as controls five mice injected with vehicle only (Figure 6A, toxicity 2). Five mice were sacrificed after the 14 day course of MI-2 administration (together with the five controls) and the other five mice were sacrificed after a 10 day washout period to assess delayed toxicity. No toxic effects or other indicators of sickness, including weight loss (Figure 6B, toxicity 2) or tissue damage (macroscopic or microscopic), were noted (Figure 6C). Brain, heart, lung, liver, kidney, bowel, spleen, thymus, and bone marrow tissues were examined. Bone marrow was normocellular with trilineage maturing hematopoiesis. Myeloid-to-erythroid ratio was 4–5:1. Megakaryocytes were normal in number and distribution. There was no fibrosis or increased number of blasts or lymphocytes. Complete peripheral blood counts, biochemistry, and liver function tests were normal (Table S2), These studies established the safety of MI-2 for use in antilymphoma efficacy studies.
Figure 6. MI-2 Is Nontoxic to Animals.
(A) Dose scheme for toxicity 1 (increasing daily dose) and toxicity 2 (same daily dose) experiments show daily and cumulative doses for each experiment.
(B) The y axis depicts mean ± SEM body weight of animals studied in both the incremental and cumulative dose schemes. Toxicity 1, p = 0.59 t test; toxicity 2, p = 0.52 t test.
(C) H&E staining for the indicated tissues in animals treated with vehicle, MI-2 25 mg/kg daily for 14 days, or MI-2 25 mg/kg daily for 14 days followed by a 10 day washout.
See also Table S2.
MI-2 Suppresses Human ABC-DLBCL Xenografts and Primary Human DLBCLs Ex Vivo
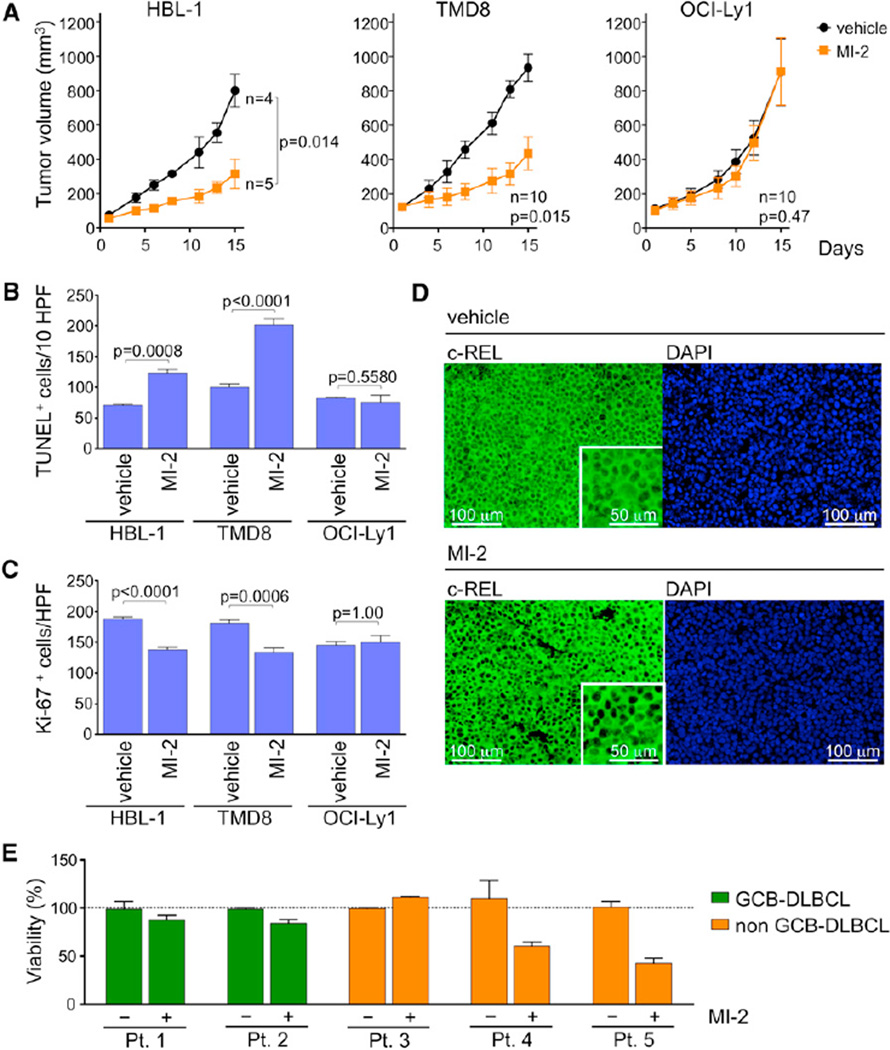
In order to determine whether MI-2 could suppress DLBCLs in vivo, we engrafted HBL-1 and TMD8 (MALT1-dependent) and OCI-Ly1 (MALT1-independent) DLBCL cells into the right flank region of nonobese diabetic/severe combined immunodeficiency (NOD-SCID) mice. Once tumors reached an average of 120 mm3 in volume, mice were randomized to receive IP injection of MI-2 25 mg/kg/day or vehicle (5% DMSO). Animals were sacrificed 24 hr after the 14th injection. Remarkably, MI-2 profoundly suppressed the growth of both the TMD8 and HBL-1 ABC-DLBCL xenografts versus vehicle, whereas it had no effect on the growth of OCI-Ly1 tumors (Figure 7A). The fact that OCI-Ly1 tumors were unaffected suggests that MI-2 activity is due to its effects on lymphoma cells rather than the host microenvironment. Histological examination using the TUNEL assay to detect apoptotic cells showed a significant increase in apoptotic cells in MI-2-treated HBL-1 and TMD8 xenografts relative to vehicle but not in OCI-Ly1 xenografts (Figures 7B and S6A). We also observed a significant decrease in proliferation as measured by Ki-67 staining in HBL-1 and TMD8 xenografts compared to vehicle, but observed no difference in OCI-Ly1 xenografts (Figures 7C and S6B). To evaluate the effect of MI-2 treatment on NF-κB signaling in xenografts, c-REL immunofluorescence was performed in paraffinized tumor sections. Consistent with data shown in Figures 4B and 4C, MI-2-treated tumors exhibited reduced c-REL nuclear protein (Figures 7D and S6C). Therefore, the MI-2 small-molecule MALT1 inhibitor specifically suppresses proliferation, survival, and NF-κB activity in ABC-DLBCLs in vivo in a lymphoma cell-autonomous manner.
Figure 7. MI-2 Suppresses ABC-DLBCLs In Vivo and Primary Human DLBCL Ex Vivo.
(A) Tumor growth curve for HBL-1, TMD8, and OCI-Ly1 xenografts in NOD-SCID mice treated with 25 mg/kg/day MI-2 or the same volume of vehicle for 14 consecutive days. Statistics, paired t test.
(B) TUNEL+ staining in histologic sections of HBL-1, TMD8, and OCI-Ly1 xenografts. The y axis represents the number of TUNEL+ cells per ten high-power fields (HPFs; n = 5 tumors/treatment, n = 3 for HBL-1). Statistics, t test.
(C) Ki-67 immunohistochemistry staining of xenografted HBL-1, TMD8, and OCI-Ly1 tumor sections. The y axis represents Ki-67+ cells per high-power field (5 HPFs/tumor for n = 5 where analyzed for each treatment, n = 3 for HBL-1). Statistics, t test.
(D) Immunohistochemical detection of c-REL in sections from HBL-1 xenografts exposed to MI-2 or vehicle. Insets are 2× digital amplification. Representative image of three HPFs analyzed for n = 3 tumors stained for each treatment.
(E) Viability assays were performed in quadruplicate in primary human DLBCL patient samples treated for 48 hr with 0.8 µM MI-2 or vehicle. The y axis represents % of viable cells normalized to vehicle. Data are presented as mean ± SEM for all panels.
See also Figure S6.
Finally, to determine whether MI-2 could also suppress primary human DLBCLs, we obtained single-cell suspensions from lymph node biopsies of five DLBCL patients for whom their GCB versus non-GCB status could be ascertained by immunohistochemistry using the Hans criteria (Hans et al., 2004), as a surrogate for GCB versus ABC classification. Lymphoma cells were isolated and exposed to 0.8 µM MI-2 or vehicle in four replicates. After 48 hr exposure, cell number and viability were determined using trypan blue. Notably, two of the non-GCB cases responded to MI-2 (p = 0.04 and 0.003 versus vehicle), whereas none of the GCBs did (Figure 7E). One of the non-GCB cases did not respond to MI-2; perhaps this case was not accurately classified by Hans’s criteria. Overall, these studies indicate that therapeutic targeting of MALT1 using the MI-2 small-molecule inhibitor has powerful suppressive effects on human ABC-DLBCL cells and warrants translation for use in clinical trials.
DISCUSSION
CBM complex signaling is constitutively active in a subset of ABC-DLBCLs due to somatic mutations of various genes leading to constitutive MALT1 signaling and NF-κB activation (Davis et al., 2010). The catalytic activity of MALT1 is well defined and involves substrate features such as peptide length and amino acid composition and position (Hachmann et al., 2012). Purified MALT1 is not very active in solution, because it is present as a monomer instead of its active dimeric form. Dimerization can be induced by high-salt concentrations, 1 M sodium citrate (Boatright et al., 2003). However, these high-salt conditions are nonphysiological and unsuitable for screening physiologically relevant small-molecule inhibitors. Use of a constitutively dimeric form of MALT1 enabled us to screen and identify potential inhibitors. Among these, MI-2 was found to be a potent, selective, and irreversible MALT1 inhibitor, analogous to protease inhibitor drugs such as telaprevir against the NS3/4A protease of hepatitis C virus (Klibanov et al., 2011), the proteasome inhibitor carfilzomib (Genin et al., 2010), and others (Powers et al., 2002). Although the peptide inhibitor Z-VRPR-FMK has been useful as a research tool, it is not suitable as a MALT1 therapeutic agent given its relatively large size, charge, and consequent lower cell permeability. Accordingly, MI-2 displayed superior activity in cell-based assays with excellent cell penetration and indeed featured high concentration within cells, and yet was still highly selective for MALT1 versus other caspases. Notably, a selective and irreversible small-molecule inhibitor of the tyrosine kinase BTK, PCI-32765 (ibrutinib), is currently under clinical development in patients with B cell non-Hodgkin’s lymphoma (Honigberg et al., 2010). Irreversibility of MI-2 may provide pharmacokinetic advantages. As ABC-DLBCLs have chronically active BCR signaling, prolonged suppression of MALT1 cleavage would likely be necessary for maximal antilymphoma activity. Using an irreversible inhibitor, activity will only return when new enzyme is synthesized. This may allow drugs to be effective at a lower plasma concentration, thus reducing dosing level and frequency, limiting the requirement for a long plasma half-life without compromising efficacy, and minimizing potential toxic effects related to prolonged exposure to circulating drugs. Indeed, our detailed studies indicated that MI-2 was nontoxic in animals. This result is consistent with the fact that MALT1 is the only paracaspase in humans and that MALT1-deficient mice are relatively healthy (Ruefli-Brasse et al., 2003; Ruland et al., 2003).
Chronic activation of the BCR pathway in ABC-DLBCL is mediated by several different mechanisms, many of them upstream of MALT1. ABC-DLBCL is addicted to this pathway and is often specifically addicted to MALT1 cleavage activity (Ferch et al., 2009; Hailfinger et al., 2009; Ngo et al., 2006). Notably, MI-2 selectively killed ABC-DLBCL cell lines with CD79A/B, CARMA1, and/or MYD88 mutations but not those occurring in proteins downstream of MALT1, including those with A20 homozygous deletion or TAK1 mutation. These findings underline the importance of targeted resequencing of recurrently mutated alleles in lymphoma for the rational deployment of targeted therapeutics. Although the full spectrum of lymphomas that can be targeted with MALT1 inhibitors is not fully clear yet, using an ex vivo system we were able to show that primary human non-GCB-DLBCL specimens are also addicted to MALT1 and are suppressed by MI-2.
As single agents are generally not curative and rapidly generate resistance (Misale et al., 2012), there is a growing interest in combinatorial-targeted therapy. Rational combination of MALT1 cleavage inhibition could include a combination with tyrosine kinase inhibitors targeting the Src family (dasatinib, saracatinib, bosutinib, and KX01), SYK (fostamatinib disodium), or BTK (PCI-32765). These drugs would likely synergize with MALT1 cleavage inhibition of NF-κB by further inhibiting BCR signaling, including mitogen-activated protein kinases and phosphatidylinositol 3-kinase (Lim et al., 2012). Protein kinase C (PKC) inhibition would also be a potentially beneficial combination, as it could further inhibit the NF-κB pathway, including those activities dependent on MALT1 but independent of its proteolytic activity. The PKC inhibitor sotrastaurin, in clinical trials for prevention of transplantation rejection and treatment of psoriasis (Manicassamy, 2009; Matz et al., 2011), has been recently shown to inhibit growth of ABC-DLBCL xenografted tumors (Naylor et al., 2011), pointing to its potential use as an antilymphoma therapy for this lymphoma subtype. ABC-DLBCLs also feature BCL6 translocation, SPI-B amplification, or PRDM1 deletion or mutation (Lenz and Staudt, 2010). BCL6 inhibitors promote apoptosis and cell-cycle arrest through release of critical checkpoint genes (Cerchietti et al., 2009, 2010a; Polo et al., 2004). Combination of MI-2 and BCL6 inhibitors would thus suppress two critical pathways in ABC-DLBCLs (BCL6 and NF-κB), potentially leading to therapeutic synergy. Taken together, the results reported here identify MI-2 as a lead compound targeting MALT1 and demonstrate the significance, safety, and efficacy of MALT1 as a therapeutic target and MI-2 as a therapeutic agent for the treatment of aggressive non-Hodgkin’s lymphomas that are both dependent on NF-κB signals and resistant to conventional chemotherapeutic regimens.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures are presented in Supplemental Experimental Procedures.
High-Throughput Screening for MALT1 Proteolytic Activity Inhibitors
Ac-LRSR-AMC was used as substrate and reactions were measured with excitation/emission wavelengths of 360/465 nm. Two time points were measured for each reaction to eliminate false positives due to compound autofluorescence. The final percent inhibition was calculated with the formula {[fluorescencetest compound (T2 − T1) − fluorescenceneg ctrl (T2 − T1)]/[fluorescencepos ctrl (T2 − T1) − fluorescenceneg ctrl (T2 − T1)]} × 100, where Z-VRPR-FMK (300 nM) was used as positive control and buffer only as negative control. The positive hits were validated in concentration-response experiments within a dose range of 0.122–62.5 µM to determine IC50 of the compounds. Activity was validated using recombinant full-length wild-type MALT1.
Growth-Inhibition Determination
Cell proliferation was determined by ATP quantification using a luminescent method (CellTiter-Glo; Promega) and trypan blue dye exclusion. Standard curves for each cell line were calculated by plotting the cell number (determined using trypan blue) against their luminescence values, and cell number was calculated accordingly. Cell viability in drug-treated cells was normalized to their respective controls (fractional viability), and results are given as 1-fractional viability. CompuSyn software (Biosoft) was used to determine GI25 and GI50 values.
Mouse Xenograft Experiments
Eight-week-old male SCID NOD.CB17-Prkdcscid/J mice were purchased from Jackson Laboratories and housed in a clean environment. Mice were subcutaneously injected with low-passage 107 human HBL-1, TMD8, or OCI-Ly1 cells in 50% Matrigel. Treatment was initiated when tumors reached an average size of 120 mm3. MI-2 was reconstituted in DMSO and stored at −80°C until used and was administered by intraperitoneal injection. Tumor volume was monitored by digital calipering three times a week and calculated using the formula (smallest diameter2 × largest diameter)/2. All procedures involving animals followed National Institutes of Health protocols and were approved by the Animal Institute Committee of the Weill Cornell Medical College of Cornell University.
Primary Cells
Patient-deidentified tissues were obtained in accordance with the guidelines and approval of the University of Navarra Institutional and Weill Cornell Medical College Review Boards. Only discarded leftover tissue after diagnosis was rendered was utilized for research, in agreement with institutional review board protocol. Patient samples were processed as previously described (Cerchietti et al., 2010b). Briefly, single-cell suspensions from lymph node biopsies were obtained by physical disruption of tissues followed by cell-density gradient separation. Cell number and viability were determined by trypan blue exclusion. Primary DLBCL cells were cultured in 96-well plates. Cells were grown in RPMI medium with 20% FBS supplemented with antibiotics for 48 hr. Cells were exposed to 0.8 µM MI-2 (1 µM for Pt.2) or control (DMSO) in quadruplicate. After 48 hr of exposure, viability was determined by using trypan blue (Sigma). All samples were normalized to their own replicate control.
Supplementary Material
Significance.
MALT1 is a unique paracaspase protein that transduces aberrant oncogenic signaling in ABC-DLBCL. This manuscript reports the development of a constitutively activated form of MALT1 that enabled a screen for small molecule inhibitors. This led to identification of MI-2, an irreversible MALT1 protease inhibitor, which is a lead compound with nanomolar activity in cell-based assays and selective activity against ABC-DLBCLs. Importantly, we show that MALT1 inhibitors kill ABC-DLBCLs in vitro and in vivo, are nontoxic to animals, and also suppress primary human non-GCB-DLBCL specimens. Hence, we demonstrate that MALT1 is a bona fide therapeutic target and provide a lead compound that forms the basis of a class of therapeutic agents for B cell lymphomas.
ACKNOWLEDGMENTS
We thank Paulus Erbel, Martin Renatus, and Nicola Hughes (Novartis Institutes for BioMedical Research, Basel, Switzerland) for mass spectrometry, NMR experiments, and advice; Beatriz Bellosillo (H. del Mar, Barcelona, Spain) and Miguel A. Piris (Hospital Marques de Valdecillas, Santander, Spain) for providing lymphoma samples; and Ermelinda Damko, Liwei Wang, Qi Qiao, and Jennifer Ishii for technical assistance. L.F. is supported by the Spanish Ministry of Science Fellowship “Sara Borrell.” C.Y. was supported by the Irvington Institute Postdoctoral Fellowship Program of the Cancer Research Institute. H.W. and A.M. are supported by Leukemia and Lymphoma Society Translational Research Program 6210-12. A.M. is a Burroughs Wellcome Clinical Translational Scientist and is supported by the Chemotherapy Foundation and the Beverly and Raymond Sackler Center for Physical and Biomedical Sciences.
Footnotes
ACCESSION NUMBERS
The Gene Expression Omnibus accession number for the expression microarray data reported in this paper is GSE40003.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures, two tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.ccr.2012.11.003.
REFERENCES
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS. A unified model for apical caspase activation. Mol. Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- Cerchietti LC, Yang SN, Shaknovich R, Hatzi K, Polo JM, Chadburn A, Dowdy SF, Melnick A. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood. 2009;113:3397–3405. doi: 10.1182/blood-2008-07-168773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, Bunting KL, Polo JM, Farès C, Arrowsmith CH, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010a;17:400–411. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchietti LC, Hatzi K, Caldas-Lopes E, Yang SN, Figueroa ME, Morin RD, Hirst M, Mendez L, Shaknovich R, Cole PA, et al. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J. Clin. Invest. 2010b;120:4569–4582. doi: 10.1172/JCI42869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, et al. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-κB inhibitor A20. Nat. Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, De Wolf-Peeters C, Hagemeijer A, Van den Berghe H, Marynen P. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood. 1999;93:3601–3609. [PubMed] [Google Scholar]
- Dierlamm J, Murga Penas EM, Bentink S, Wessendorf S, Berger H, Hummel M, Klapper W, Lenze D, Rosenwald A, Haralambieva E, et al. Deutsche Krebshilfe Network Project “Molecular Mechanisms in Malignant Lymphomas”. Gain of chromosome region 18q21 including the MALT1 gene is associated with the activated B-cell-like gene expression subtype and increased BCL2 gene dosage and protein expression in diffuse large B-cell lymphoma. Haematologica. 2008;93:688–696. doi: 10.3324/haematol.12057. [DOI] [PubMed] [Google Scholar]
- Farinha P, Gascoyne RD. Molecular pathogenesis of mucosa-associated lymphoid tissue lymphoma. J. Clin. Oncol. 2005;23:6370–6378. doi: 10.1200/JCO.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Ferch U, zum Büschenfelde CM, Gewies A, Wegener E, Rauser S, Peschel C, Krappmann D, Ruland J. MALT1 directs B cell receptor-induced canonical nuclear factor-κB signaling selectively to the c-Rel subunit. Nat. Immunol. 2007;8:984–991. doi: 10.1038/ni1493. [DOI] [PubMed] [Google Scholar]
- Ferch U, Kloo B, Gewies A, Pfänder V, Düwel M, Peschel C, Krappmann D, Ruland J. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 2009;206:2313–2320. doi: 10.1084/jem.20091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin E, Reboud-Ravaux M, Vidal J. Proteasome inhibitors: recent advances and new perspectives in medicinal chemistry. Curr. Top. Med. Chem. 2010;10:232–256. doi: 10.2174/156802610790725515. [DOI] [PubMed] [Google Scholar]
- Gross O, Grupp C, Steinberg C, Zimmermann S, Strasser D, Hannesschläger N, Reindl W, Jonsson H, Huo H, Littman DR, et al. Multiple ITAM-coupled NK-cell receptors engage the Bcl10/Malt1 complex via Carma1 for NF-κB and MAPK activation to selectively control cytokine production. Blood. 2008;112:2421–2428. doi: 10.1182/blood-2007-11-123513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachmann J, Snipas SJ, van Raam BJ, Cancino EM, Houlihan EJ, Poreba M, Kasperkiewicz P, Drag M, Salvesen GS. Mechanism and specificity of the human paracaspase MALT1. Biochem. J. 2012;443:287–295. doi: 10.1042/BJ20120035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailfinger S, Lenz G, Ngo V, Posvitz-Fejfar A, Rebeaud F, Guzzardi M, Penas EM, Dierlamm J, Chan WC, Staudt LM, Thome M. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA. 2009;106:19946–19951. doi: 10.1073/pnas.0907511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailfinger S, Nogai H, Pelzer C, Jaworski M, Cabalzar K, Charton JE, Guzzardi M, Décaillet C, Grau M, Dörken B, et al. Malt1-dependent RelB cleavage promotes canonical NF-κB activation in lymphocytes and lymphoma cell lines. Proc. Natl. Acad. Sci. USA. 2011;108:14596–14601. doi: 10.1073/pnas.1105020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Müller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- Honigberg LA, Smith AM, Sirisawad M, Verner E, Loury D, Chang B, Li S, Pan Z, Thamm DH, Miller RA, Buggy JJ. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc. Natl. Acad. Sci. USA. 2010;107:13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma K, Tsuzuki S, Nakagawa M, Tagawa H, Nakamura S, Morishima Y, Seto M. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009;114:2467–2475. doi: 10.1182/blood-2008-12-194852. [DOI] [PubMed] [Google Scholar]
- Klibanov OM, Williams SH, Smith LS, Olin JL, Vickery SB. Telaprevir: a novel NS3/4 protease inhibitor for the treatment of hepatitis C. Pharmacotherapy. 2011;31:951–974. doi: 10.1592/phco.31.10.951. [DOI] [PubMed] [Google Scholar]
- Lenz G, Staudt LM. Aggressive lymphomas. N. Engl. J. Med. 2010;362:1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008a;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl. Acad. Sci. USA. 2008b;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Yang Y, Staudt LM. Pathogenetic importance and therapeutic implications of NF-κB in lymphoid malignancies. Immunol. Rev. 2012;246:359–378. doi: 10.1111/j.1600-065X.2012.01105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas PC, Yonezumi M, Inohara N, McAllister-Lucas LM, Abazeed ME, Chen FF, Yamaoka S, Seto M, Nunez G. Bcl10 and MALT1, independent targets of chromosomal translocation in MALT lymphoma, cooperate in a novel NF-κB signaling pathway. J. Biol. Chem. 2001;276:19012–19019. doi: 10.1074/jbc.M009984200. [DOI] [PubMed] [Google Scholar]
- Manicassamy S. Sotrastaurin, a protein kinase C inhibitor for the prevention of transplant rejection and treatment of psoriasis. Curr. Opin. Investig. Drugs. 2009;10:1225–1235. [PubMed] [Google Scholar]
- Matz M, Naik M, Mashreghi MF, Glander P, Neumayer HH, Budde K. Evaluation of the novel protein kinase C inhibitor sotrastaurin as immunosuppressive therapy after renal transplantation. Expert Opin. Drug Metab. Toxicol. 2011;7:103–113. doi: 10.1517/17425255.2011.540238. [DOI] [PubMed] [Google Scholar]
- Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor TL, Tang H, Ratsch BA, Enns A, Loo A, Chen L, Lenz P, Waters NJ, Schuler W, Dörken B, et al. Protein kinase C inhibitor sotrastaurin selectively inhibits the growth of CD79 mutant diffuse large B-cell lymphomas. Cancer Res. 2011;71:2643–2653. doi: 10.1158/0008-5472.CAN-10-2525. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Dell’Oso T, Ranuncolo SM, Cerchietti L, Beck D, Da Silva GF, Prive GG, Licht JD, Melnick A. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat. Med. 2004;10:1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- Pop C, Timmer J, Sperandio S, Salvesen GS. The apoptosome activates caspase-9 by dimerization. Mol. Cell. 2006;22:269–275. doi: 10.1016/j.molcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- Rebeaud F, Hailfinger S, Posevitz-Fejfar A, Tapernoux M, Moser R, Rueda D, Gaide O, Guzzardi M, Iancu EM, Rufer N, et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat. Immunol. 2008;9:272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, et al. Lymphoma/Leukemia Molecular Profiling Project. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, Chan WC, Zhao T, Haioun C, Greiner TC, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J. Exp. Med. 2003;198:851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruefli-Brasse AA, French DM, Dixit VM. Regulation of NF-κB-dependent lymphocyte activation and development by paracaspase. Science. 2003;302:1581–1584. doi: 10.1126/science.1090769. [DOI] [PubMed] [Google Scholar]
- Ruland J, Duncan GS, Wakeham A, Mak TW. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity. 2003;19:749–758. doi: 10.1016/s1074-7613(03)00293-0. [DOI] [PubMed] [Google Scholar]
- Sanchez-Izquierdo D, Buchonnet G, Siebert R, Gascoyne RD, Climent J, Karran L, Marin M, Blesa D, Horsman D, Rosenwald A, et al. MALT1 is deregulated by both chromosomal translocation and amplification in B-cell non-Hodgkin lymphoma. Blood. 2003;101:4539–4546. doi: 10.1182/blood-2002-10-3236. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J. Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Staal J, Driege Y, Bekaert T, Demeyer A, Muyllaert D, Van Damme P, Gevaert K, Beyaert R. T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. EMBO J. 2011;30:1742–1752. doi: 10.1038/emboj.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel B, Lamprecht A, Dierlamm J, Cerroni L, Stolte M, Ott G, Raderer M, Chott A. T(14;18)(q32;q21) involving IGH and MALT1 is a frequent chromosomal aberration in MALT lymphoma. Blood. 2003;101:2335–2339. doi: 10.1182/blood-2002-09-2963. [DOI] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. Lyon: IARC Press; 2008. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues. [Google Scholar]
- Vicente-Dueñas C, Fontán L, Gonzalez-Herrero I, Romero-Camarero I, Segura V, Aznar MA, Alonso-Escudero E, Campos-Sanchez E, Ruiz-Roca L, Barajas-Diego M, et al. Expression of MALT1 oncogene in hematopoietic stem/progenitor cells recapitulates the pathogenesis of human lymphoma in mice. Proc. Natl. Acad. Sci. USA. 2012;109:10534–10539. doi: 10.1073/pnas.1204127109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker NP, Talanian RV, Brady KD, Dang LC, Bump NJ, Ferenz CR, Franklin S, Ghayur T, Hackett MC, Hammill LD, et al. Crystal structure of the cysteine protease interleukin-1β-converting enzyme: a (p20/p10)2 homodimer. Cell. 1994;78:343–352. doi: 10.1016/0092-8674(94)90303-4. [DOI] [PubMed] [Google Scholar]
- Wiesmann C, Leder L, Blank J, Bernardi A, Melkko S, Decock A, D’Arcy A, Villard F, Erbel P, Hughes N, et al. Structural determinants of MALT1 protease activity. J. Mol. Biol. 2012;419:4–21. doi: 10.1016/j.jmb.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Park HH, Chung JY, Lin SC, Lo YC, da Graca LS, Jiang X, Wu H. Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol. Cell. 2006;22:259–268. doi: 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Jeffrey PD, Ha JY, Yang X, Shi Y. Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region. Proc. Natl. Acad. Sci. USA. 2011;108:21004–21009. doi: 10.1073/pnas.1111708108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.