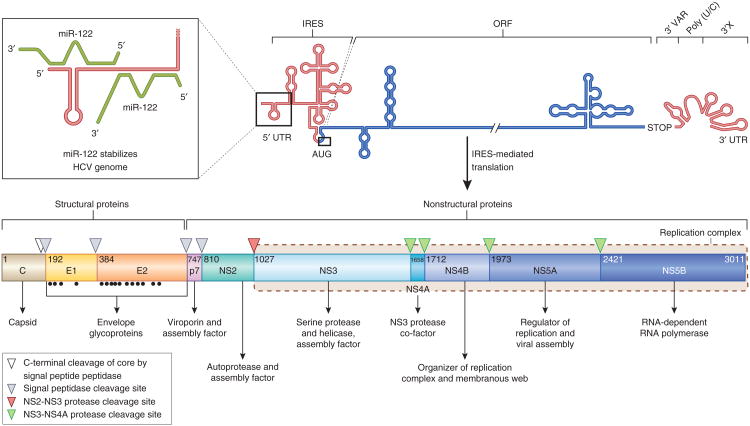
Figure 2.
The HCV genome and polyprotein processing. The HCV RNA genome (top) contains one long ORF (blue) flanked by 5′ and 3′ UTRs (red). The binding of two copies of miR-122 (green) to the 5′ UTR is highlighted in the inset. IRES-mediated translation of the ORF leads to a polyprotein (bottom) that is co- and post-translationally processed into ten viral proteins. The maturation process of the core protein involves a cellular signa peptide peptidase cleavage of a C-terminal signal peptide (white triangle) and cleavage from E1 by the cellular signal peptidase123, which also cleaves E1, E2 and p7 from the polyprotein (gray triangles). In an autocleavage mechanism requiring two identical molecules to make up the composite active site, the NS2-NS3 protease cleaves itself (red triangle)72. The NS3 protease located in the first one-third of NS3 (ref. 165), assisted by its membrane-bound cofactor, NS4A166, cleaves the remaining proteins NS3, NS4A, NS4B, NS5A and NS5B (green triangles). Glycosylation of the envelope proteins (black dots) and the functions of the individual HCV proteins are indicated.