Figure 4.
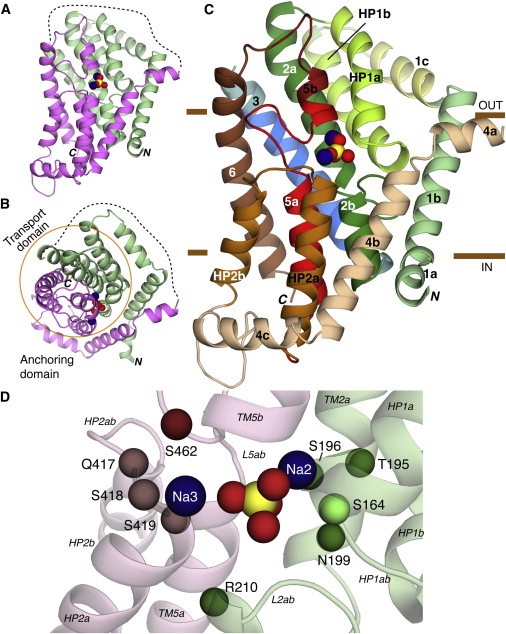
Structural model of human NaPi-IIa based on VcINDY. (A–C) Overview of the predicted fold of the NaPi-IIa model represented by cartoon helices. Two sodium ions (blue) and Pi (yellow, red) are shown as spheres. The model is viewed (A and C) from within the plane of the membrane, or (B) from the extracellular side. (A and B) Location of the structural repeats RU1 (green) and RU2 (pink), and the connecting long extracellular loop (dashed lines). The transport domain is highlighted by an orange circle, in B. In C, individual helices are colored according to Fig. 3A. Regions for which experimental data relating to the topology are highlighted: residues accessible to the extracellular side (yellow) or buried (blue) in helix 1c and 3, respectively; and residues expected to be helical in TM5a, TM5b, and the 5ab loop (red). The approximate extents of the membrane are indicated by brown bars. (D) The predicted binding site region in the model of NaPi-IIa. The Cα-atoms of conserved and polar residues close to the putative binding site are highlighted, as are bound Pi and sodium ions (spheres). Residues N199 and S462 have previously been implicated in substrate binding. Residues S164, T195, S196, R210, Q417, S418, S419 are investigated in this study. To see this figure in color, go online.
