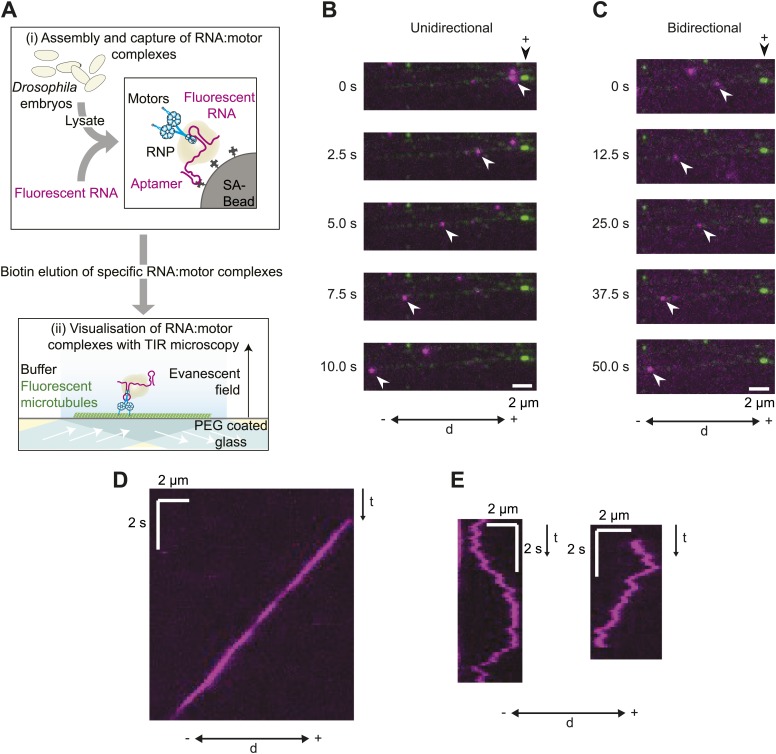
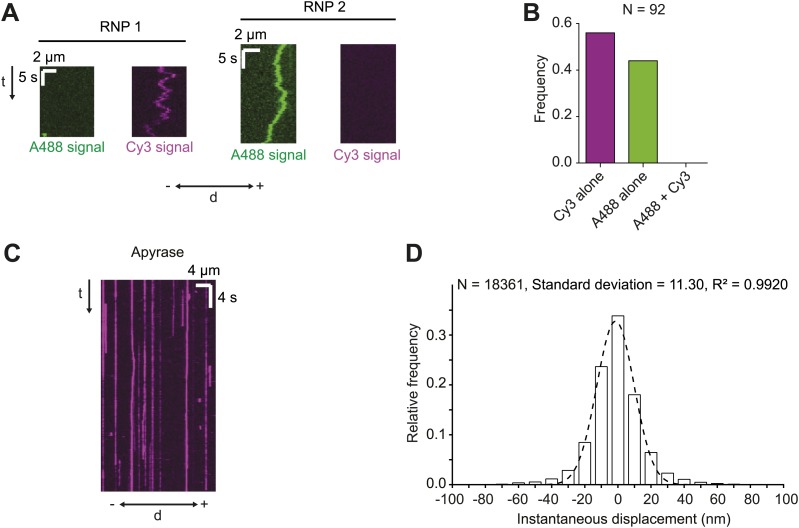
(
A and
B) Motile RNPs assembled in the presence of equimolar Alexa488 (A488)-labelled
h and Cy3-labelled
h contained only A488 or Cy3 dyes, but never both. Thus, single fluorescent
h molecules were present in each RNP. Kymographs of two examples of RNPs (
A) and overall quantification from the experimental series (
B) are shown. d, distance; −, minus end; +, plus end; N, number of motile RNPs analysed. For the experiments in
A and
B the Alexa488-labelled RNAs and Cy3-labelled RNAs were mixed and incubated with the streptavidin bead matrix before the addition of extract. (
C) Representative kymograph of
h RNPs in the presence of 20 U·ml
−1 apyrase (which is used to deplete ATP and ADP from the chamber [
Higuchi et al., 1997;
Ma and Taylor, 1997]). Note that RNPs appear static. (
D) Distribution of instantaneous frame-to-frame displacements generated from automatic tracking of
h RNPs on microtubules in the presence of 20 U·ml
−1 apyrase. In accordance with previous studies (
Hendricks et al., 2010), our tracking accuracy was defined as one standard deviation of these displacements. Dashed line shows Gaussian fit from which standard deviation was calculated. R
2, goodness of fit; N, total number of individual displacements analysed (from 15 RNPs).