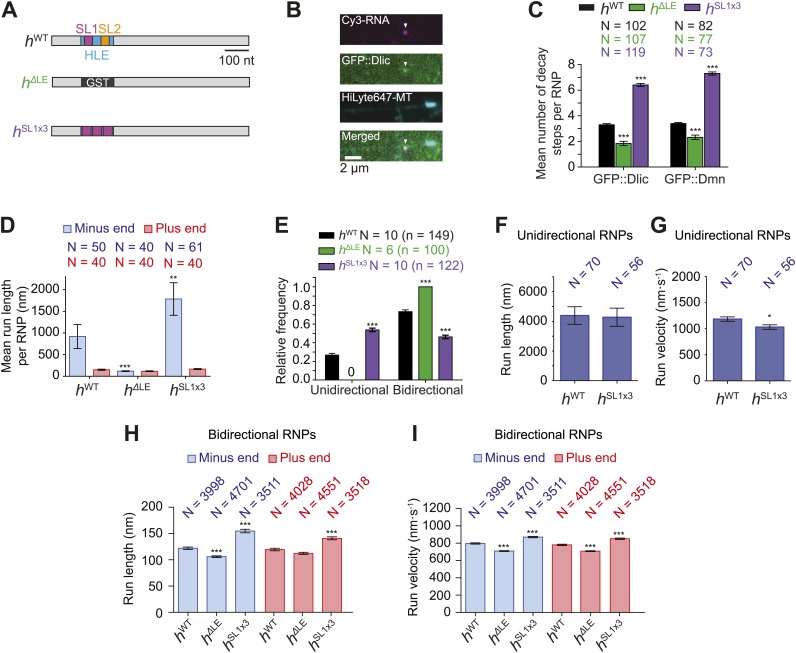
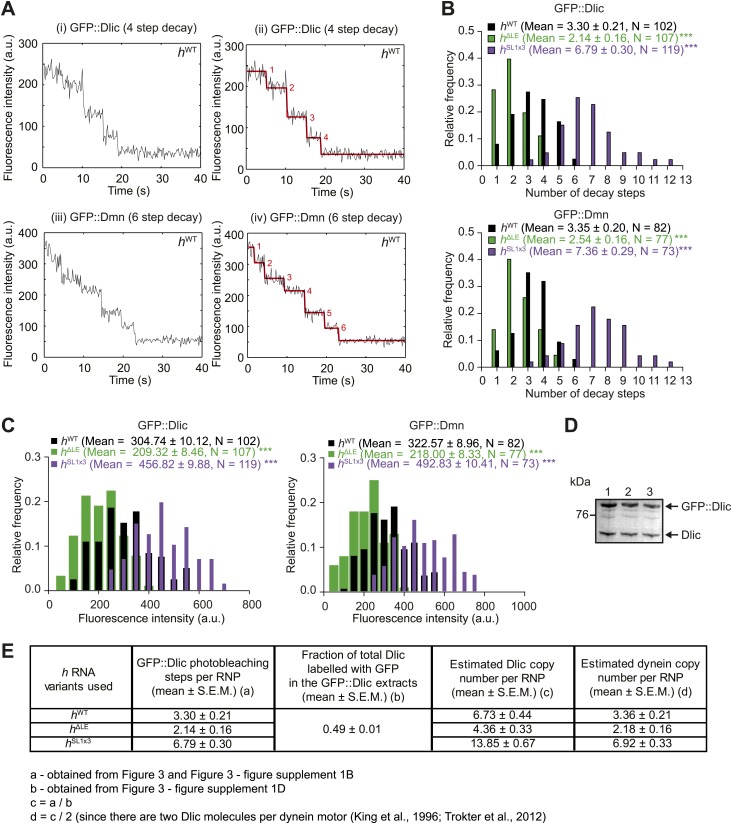
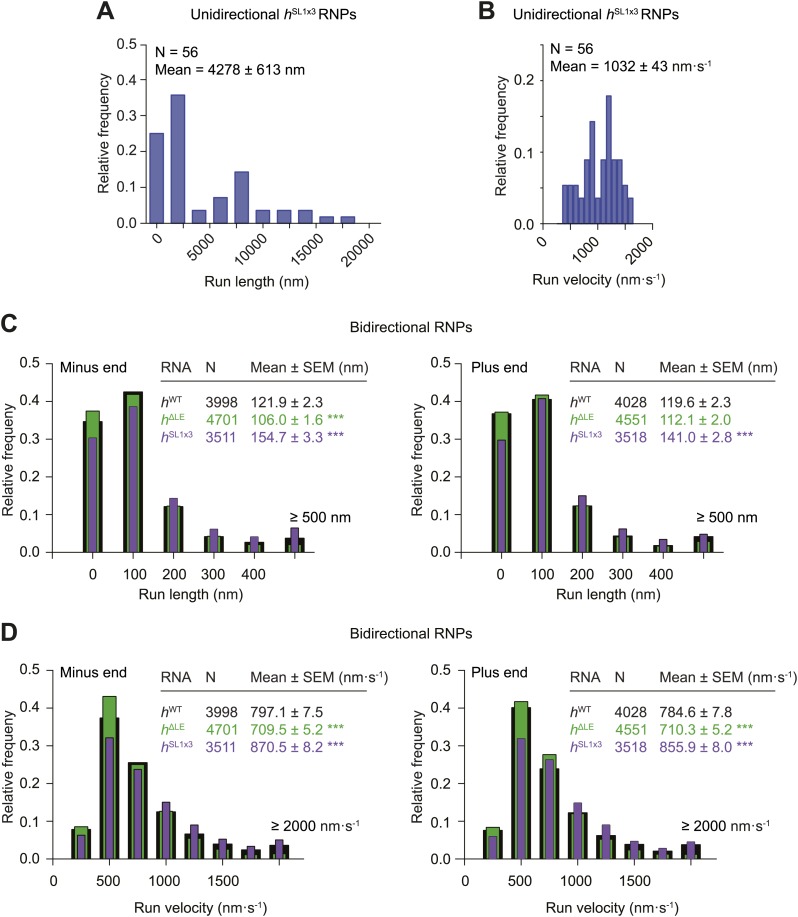
Figure 3. Manipulating the copy number of native dynein-dynactin complexes on individual RNPs.
(A) Schematic of h RNA variants fused to streptavidin aptamers and used in this study. hΔLE and hSL1x3 have replacements of the 124 nt HLE region with, respectively, a heterologous sequence from the Glutathione-S-transferase (GST) gene and three copies of stem-loop 1 (SL1) separated by short spacers. (B) Example of an RNP in vitro containing Cy3-h RNA (magenta) and GFP::Dlic (green), which is immobilised on a polarity-marked microtubule (cyan) by the omission of ATP. (C) Stepwise GFP photobleaching analysis of RNPs assembled on h RNA variants. Note significant relative changes in the copy number of Dlic and Dmn upon increasing or decreasing copy number of SL1. N, number of photobleaching traces analysed. See Figure 3—figure supplement 1B for distribution of values. (D) Mean run length per RNP for each RNA species. N, number of RNPs analysed. (E) Mean proportion of motile RNPs per imaging chamber that are unidirectional or bidirectional. No unidirectional RNPs were observed for hΔLE. N, number of chambers analysed; n, total number of RNPs. (F and G) Mean length (F) and velocity (G) of individual runs of unidirectional RNPs. N, number of runs (from 25 and 20 RNPs for hWT and hSL1x3, respectively [many RNPs have more than one run due to interruptions of bouts of minus end-directed motility by short-lived pauses]). (H and I) Mean length (H) and velocity (I) of individual runs of bidirectional RNPs. N, number of runs (from 40 RNPs each for hWT, hΔLE, and hSL1x3). In D–I, all experiments were performed in 2.5 mM ATP; error bars represent SEM. ***p<0.001; **p<0.01; *p<0.05, compared to hWT values for the same parameter (Mann–Whitney non-parametric t test).