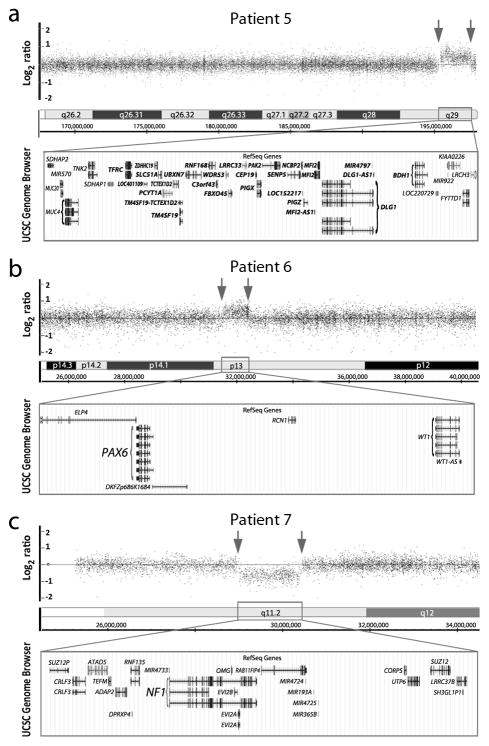
Figure 2.
Likely pathogenic copy number variants involving regions with a known role in human disease. (a–c) Affymetrix Genome-Wide Human SNP Array 6.0 data for Patients 5–7; breakpoints are indicated with arrows. The UCSC Genome Browser (http://genome.ucsc.edu) view of the affected region indicating the positions of genes for each region (GRCh37/hg19 assembly). (a) 3q29 duplication in Patient 5; genes located within the minimum critical internal (~1.58 Mb) for 3q29 microduplication syndrome as defined by Goobie et al. 2008 (19) are indicated in bold font. (b) 11p13 duplication including PAX6 in Patient 6. (c) 17q11 deletion including NF1 in Patient 7.