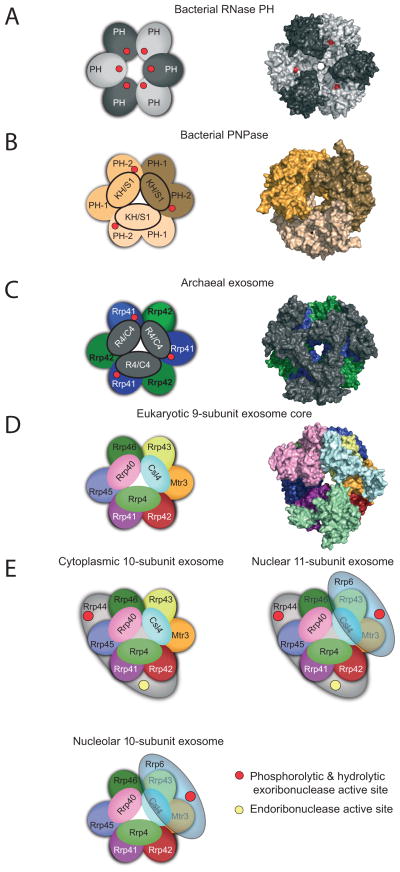
Figure 2. Conserved architecture of exosome core from bacteria, archaea, and eukaryotes.
Structures of complexes reveal a six-component ring architecture with or without phosphorolytic active sites (shown as red dots). The left panel shows the cartoon representation, and the right panel shows the x-ray structure surface representation. A) RNase PH. The Aquifex aeolicus RNase PH structure (PDB ID = 1UDN) forms a homohexamer of PH subunits (colored dark grey and light grey). B) PNPase. The S. antibioticus PNPase structure (PDB ID = 1E3P) forms a homotrimer. Each of PNPase protomers are colored differently: light yellow, dark yellow, and light brown to emphasize the homotrimer of RNase PH 1-like (PH 1) and RNase PH 2-like (PH 2) domains. C) Archaeal exosome. The S. solfataricus archaeal exosome (PDB ID = 2JE6) is shown with Rrp41 subunits (blue) and Rrp42 subunits (green). Rrp41 and Rrp42 form a heterodimer; the resulting heterodimer can trimerize, culminating in a six-component ring. Either Csl4 or Rrp4 (shown in grey) form a trimeric cap above the ring. The surface representation of the structure depicts the Rrp4-bound form. D) Eukaryotic exosome: 9-subunit core. Human subunits are labeled and color-coded and include the PH-like ring subunits Mtr3 (orange), Rrp42 (red), Rrp41 (purple), Rrp45 (blue), Rrp46 (green) and Rrp43 (yellow); the S1/KH domain proteins Csl4 (light blue), Rrp4 (green) and Rrp40 (pink). E) Cytoplasmic, nuclear and nucleolar exosome architectures. The non-catalytic exosome core interacts with additional hydrolytic enzymes to form: the 10 subunit cytoplasmic exosome (panel 1, Exo9 + Rrp44), the 11 subunit nuclear exosome (panel 2, Exo9 + Rrp44 + Rrp6), and the 10 subunit nucleolar exosome (panel 3, Exo9 + Rrp6). The S1/KH protein ring is shown on the top of the PH-like ring with Rrp44 shown below the PH-like ring to reflect structural models of the complex. Rrp6 is shown on the other side of the complex below the PH-like ring, although there is no definitive structural data for this complex. The exoribonuclease active sites are depicted with red circles in Rrp44 and Rrp6, and the endoribonuclease active site of Rrp44 is shown as a yellow circle. Graphics generated with Pymol [59].