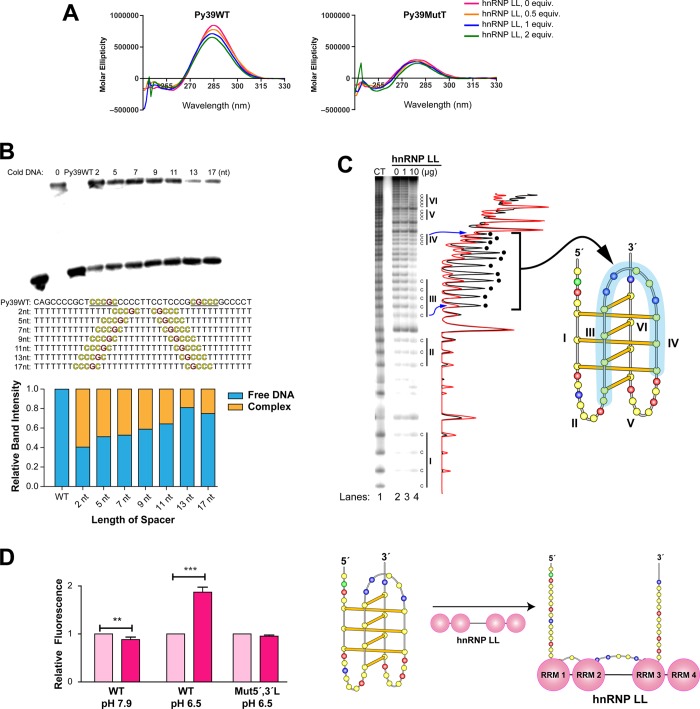
Figure 4.
EMSA, Br2 footprinting, and FRET show that hnRNP LL unfolds the BCL2 i-motif after binding. (A) CD analysis shows that binding of hnRNP LL produces a conformational change in the i-motif. hnRNP LL was preincubated with Py39WT or Py39MutT at pH 6.5 for 5 min at room temperature before measuring the CD. (B) Competition EMSA showing that 13 nt is the optimal length between two hnRNP LL binding sites for the binding of hnRNP LL. All oligomers are 39-mers. Competition EMSA experiments were conducted in a binding buffer (pH 6.8) for 20 min of preincubation of 250 nM of cold oligomers with hnRNP LL and subsequent 5 min incubation of end-labeled Py39WT. This represents about a 150 molar excess of cold DNA to labeled i-motif. The histogram below the gel shows the relative binding intensity from the EMSA gel. (C) Bromine footprinting of the BCL2 i-motif and hnRNP LL complex showing the conformational change of Py39WT induced by hnRNP LL. Py39WT and hnRNP LL were incubated for 5 min at room temperature, and bromine generated in situ was added for 30 min. Black and red plots are 0 and 10 μg of hnRNP LL, respectively. The peaks with the black dots correspond to those where maximum inhibition occurs and include C runs II and IV and the central loop. The right panel shows the folding pattern of the BCL2 i-motif with that region protected from Br2 cleavage shown in the blue shading. Experimental conditions are described in the Methods section. (D) FRET experiments showing i-motif-specific unfolding activity by hnRNP LL. FAM/TAMRA dual-labeled probes were incubated at pH 6.5 or 7.9 with hnRNP LL at room temperature for 5 min, and then fluorescence intensity was measured at 495 nm (Ex.)/528 (Em.). Right panel shows the unfolding of the i-motif consistent with the fluorescence enhancement seen in the left panel (WT at pH 6.5). P values (**P < 0.01, ***P < 0.001) were determined by t-test analysis.