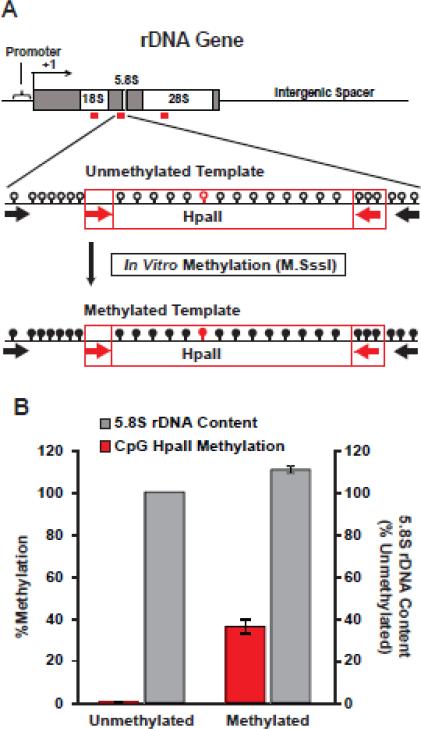
Figure 6. Effects of template methylation on qPCR efficiency.
A, PCR on genomic DNA was used to produce an unmethylated DNA fragment including a template for the 5.8S qPCR amplicon that revealed rDNA instability in DLB (red box, qPCR primers indicated by red arrows, PCR primers indicated by black arrows). DNA was then in vitro methylated with M.SssI DNA methyltransferase that indiscriminately targets all CpG sites. Note presence of multiple CpGs in the 5.8S amplicon (opened and filled circles correspond to CpGs or methyl-CpGs, respectively; one of the CpGs is a part of an HpaII site). B, Methylation of a CpG that was part of an HpaII site was confirmed using HpaII-qPCR assay. When methylated DNA fragment was used as a template for 5.8S qPCR, rDNA content was overestimated by just 10% as compared to unmethylated template. Hence, differential methylation of rDNA templates of the qPCR amplicons is unlikely to account for DLB-associated changes in rDNA content. Data represent two independent experiments; error bars are SDs.