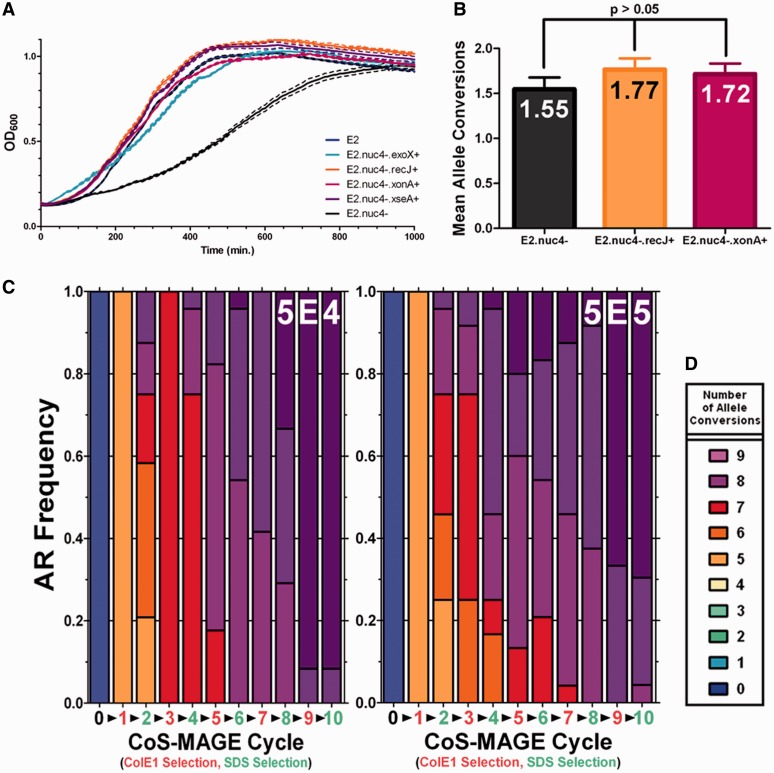
Figure 3.
tolQRA duplication enables stable CoS-MAGE cycling. (A). To probe the post-recombination growth phenotype of Nuc5−-based strains, we individually reverted each of the four inactivated nucleases: exoX+ (cyan); recJ+ (orange); xonA+ (red); xseA+ (purple). As controls, we included EcNR2 (Nuc5+, blue) and EcNR2.Nuc5− (black). To study the poor post-recombination recovery phenotype associated with EcNR2.Nuc5−, we recombined these six strains with a 5.2 µM multiplexed oligo pool, then monitored growth post-recombination. (B). To understand whether nuclease reversion results in inferior CoS-MAGE performance to Nuc5−, we tested Nuc5−, the recJ reversion (recJ+) and the xonA reversion (xonA+) strains in a single cycle of CoS-MAGE. The mascPCR data are presented as Mean Allele Conversion ± SD. Statistical analysis (Kruskal–Wallis ANOVA) revealed that the means were not statistically significantly different (P > 0.05). Moving forward, we implemented the recJ reversion in EcM2.0 (EcNR2.dnaG_Q576A.xseA-.exoX-.xonA-.1255700::tolQRA). (C and D). EcM2.0 was subjected to continuous CoS-MAGE cycling of Oligo Set 1 (22,23) using the endogenous tolCWT. We inoculated selections (SDS) using 5 × 106 cells/well, and counter-selections using 5 × 104 cells/well (C) or 5 × 105 cells/well (D). After each respective selection, clones were plated and screened for allele conversions at the 10 loci of interest using mascPCR.