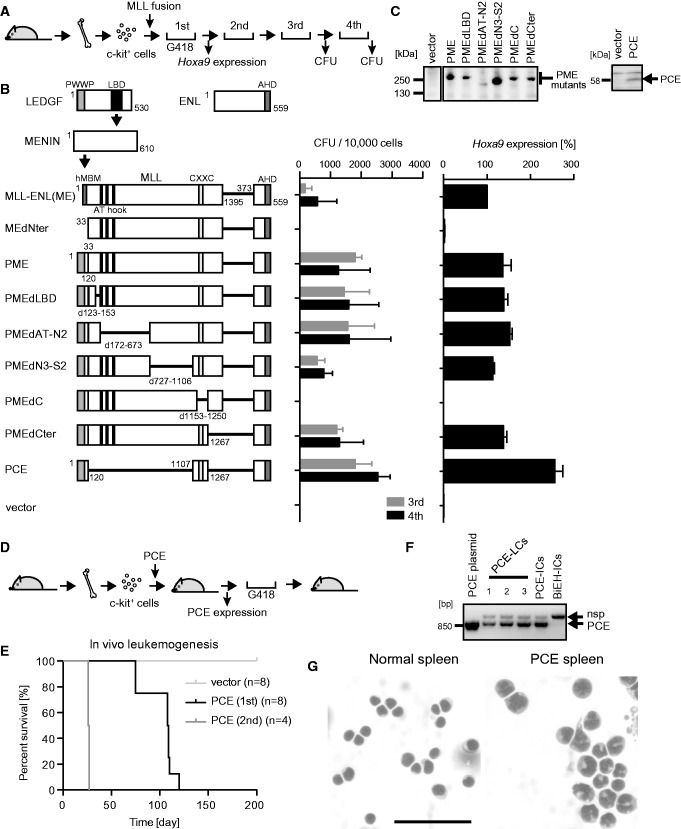
Figure 1.
Major functional domains required for leukemogenesis by MLL-ENL. (A) Experimental scheme for the myeloid progenitor transformation assay. (B) Transforming ability of various MLL-ENL (ME) mutants. The schematic structures of LEDGF, MENIN, ENL and various ME mutants are shown, with the key functional modules indicated (left). P: the PWWP domain. C: the CXXC domain. E: the ENL portion included in MLL-ENL. The numbers of colony-forming units (CFUs) at the third and fourth rounds of replating are shown with error bars (SD of >3 independent experiments) (middle). Hoxa9 expression is presented relative to the value of ME (arbitrarily set at 100%) with error bars (SD of triplicate PCRs) (right). (C) Protein expression of the PWWP-MLL-ENL (PME) and PCE mutants in the packaging cells. The PME and PCE proteins were visualized using the anti-ENL antibody. Molecular standards are shown on the left. (D) Experimental scheme for the in vivo leukemogenesis assay. (E) Survival of the transplanted animals in the in vivo leukemogenesis assay. Light gray, vector; black, first PCE transplantation; dark gray, second PCE transplantation. n, number of animals analyzed. (F) Expression of PCE in the PCE-induced leukemia cells (PCE-LCs). Leukemic cells derived from moribund mice were cultured ex vivo in the presence of G418 and analyzed by RT-PCR. The PCE plasmid, PCE-immortalized cells (PCE-ICs) and Bcl2/E2A-HLF-immortalized cells (BiEH-ICs) were included for comparison. nsp, non-specific band. (G) Morphology of PCE-LCs in the spleen. The splenic cells harvested from a PCE-leukemic mouse were stained by the May-Grunwald/Giemsa staining method. Normal spleen is included for comparison. Scale bar, 50 µm.