Figure 3.
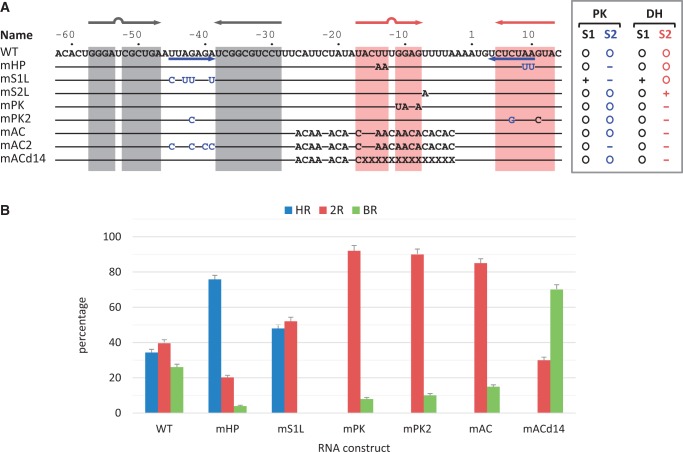
RPSOutr and the mutants. (A) List of sequences for the wild-type (WT) and mutants. The regions forming the stems of Hairpin 1 and Hairpin 2 of the double hairpin are highlighted in grey and red, respectively. Sequences forming Stem 2 of the pseudoknot are indicated by blue arrows. In the mutants, only the mutated nucleotides are shown; coloured in blue are those originally involved in pseudoknot base paring. X denotes deletion. The enclosed list to the right indicates stability changes relative to the wild-type in Stem 1 (S1) and Stem 2 (S2) of the pseudoknot (PK) and double-hairpin (DH) conformations. (B) Relative occurrence of unfolding patterns for each construct. Errors are estimated from N/√N, where N is the data number.