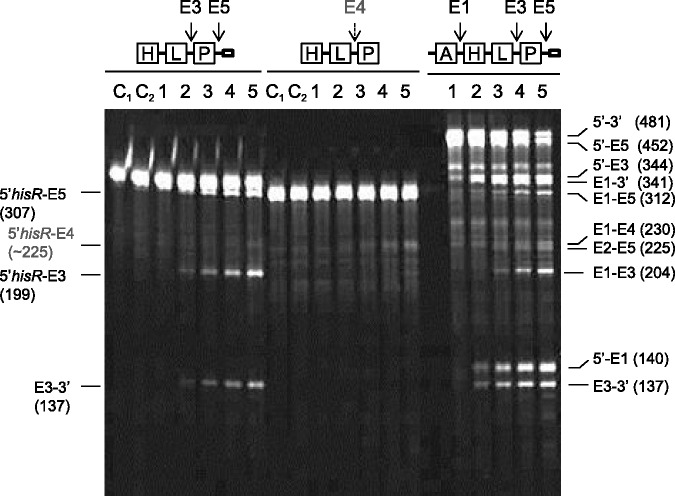
Figure 2.
The requirements for direct-entry cleavage at E3. Truncated versions of the argX-hisR-leuT-proM precursor were generated by shortening the template used for in vitro transcription. All of the transcripts had 5′-triphosphorylated ends and were incubated with T170V. The boundaries of each of the transcripts and the positions at which they were cleaved are shown schematically at the top of each panel. The cleavage of the transcripts was assayed as Figure 1. The enzyme and initial substrate concentrations at the start of each reaction were 7 and 250 nM, respectively. Lanes 1–5 contain samples taken 0, 5, 15, 30 and 60 min after mixing substrate and enzyme. Lanes C1 and C2 correspond to substrate incubated without enzyme for 0 and 60 min. The RNA was stained using SYBR® Gold stain (Life Technologies). The identities and sizes of the species produced by cleavage of the precursor starting at the 5′ end of hisR tRNA are indicated on the left of the panel. The identities and sizes of the products of cleaving full-length transcript are indicated on the right of the panel, as Figure 1.