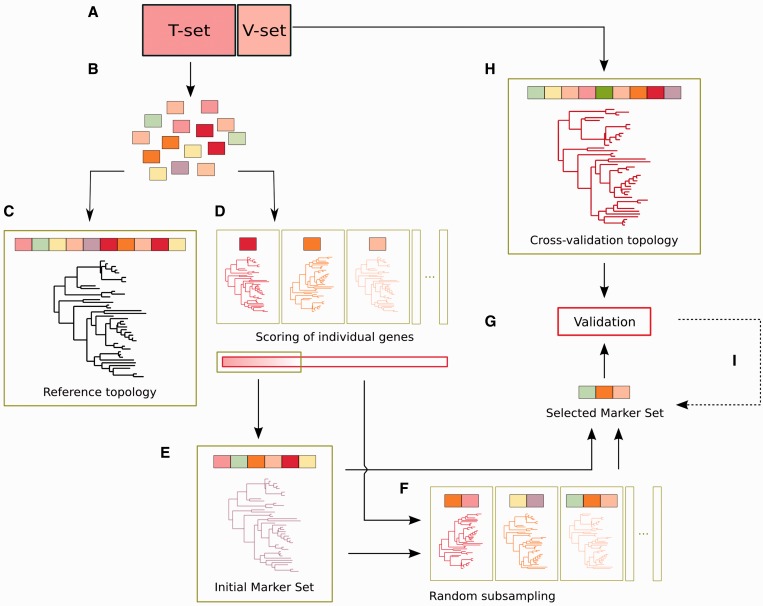
Figure 1.
Schematic representation of the proposed marker selection pipeline, including the training and the validation steps. (A) The complete set of available genomes is divided into a training and validation set. (B) For the validation set, orthologous groups of genes present in single copy in all the species are selected and aligned. (C) A phylogeny reconstructed from the analysis of the concatenated alignments is considered as the reference topology. (D) Individual alignments are also used to build individual gene phylogenies. The similarity of these phylogenies to the reference topology is used to score the phylogenetic informativeness of the genes, which are ranked accordingly. (E) Top-scoring genes are concatenated sequentially until the resulting alignment yields a phylogeny identical (or sufficiently similar according to a given similarity threshold) to the reference topology. The gene families in such a concatenated alignment constitute the ‘initial marker set’, which is sufficient to obtain the desired level of resolution; as indicated, it is possible to move directly to the validation phase or to optimize the markers set size. (F) Smaller sets of marker genes with resolving power equal to the initial marker set are searched for iteratively by random subsampling of available markers either from the whole set or from the current set of markers, and evaluation of the resulting phylogenies. The process is finished when a sufficiently small marker set with sufficiently high resolution power is found or, alternatively, when the full combinatorial space has been explored. (G) Selected marker gene sets are validated against reference topologies that include species of the validation set. (H and I) Alternative sets of marker genes can be tested if previously selected ones fail during the validation phase.