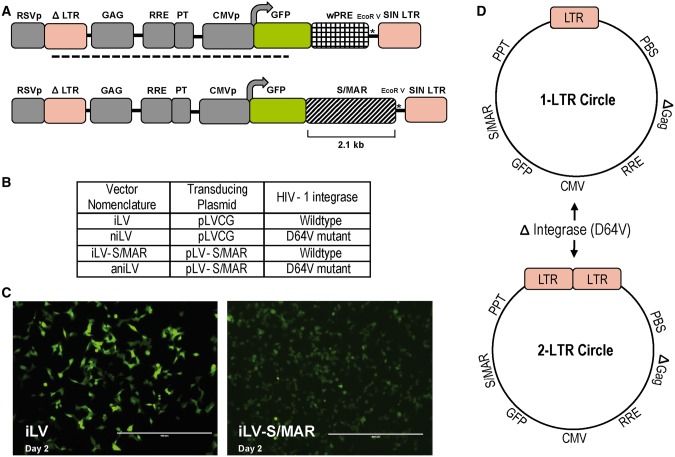
Figure 1.
Effect of wPRE replacement with S/MAR sequence on transgene expression and production of non-integrating vector episomes. (A) Diagram of HIV-1-based self-inactivating LV pLVCG with wPRE cloned within the transgene expression cassette pLVCG harboring human S/MAR from the ß-interferon gene cluster. The GFP-wPRE fragment was released by digestion with XbaI and EcoRI to replace with the S/MAR fragment. The dotted line corresponds to the DIG-labeled probe binding site. S/MAR is cloned at the transcriptionally active region between GFP reporter ORF and 3′LTR after replacing the wPRE fragment in LVCG plasmid. (B) Four LVs classified on integration status and S/MAR presence were used in the study. iLV: integrating lentiviral vector harboring wPRE sequence, niLV: non-integrating lentiviral vector harboring wPRE, iLV-S/MAR: integrating lentiviral vector harboring S/MAR sequence, aniLV: anchoring non-integrating LV harboring S/MAR sequence. (C) Microscopy images of GFP expression from iLV and iLV-S/MAR-transduced 293T cells at the day-2 time point. Integrating vector stocks were produced by using the helper plasmids pLP-1, pLP-2 and pLP/VSVG. Non-integrating vector stocks were produced by using the helper plasmids pCD/NL2 ΔInt, pLP-2 and pLP/VSVG. (D) Diagram representing the LTR circle formation due to abortive integration of niLVs. The class I integrase mutant used to produce niLVs is a D64V mutant that limits IN function regarding cleavage and integration but maintains normal levels of viral DNA. The niLVs are produced by using the helper plasmids pCD/NL2 ΔInt, pLP-2 and pLP/VSVG. After transduction, the abortive integration event of the vector genome results the formation of reverse-transcribed circular DNA vector genomes with one or two LTRs. All LTR sequences used in this study are promoterless self-inactivating (SIN) vector sequences.